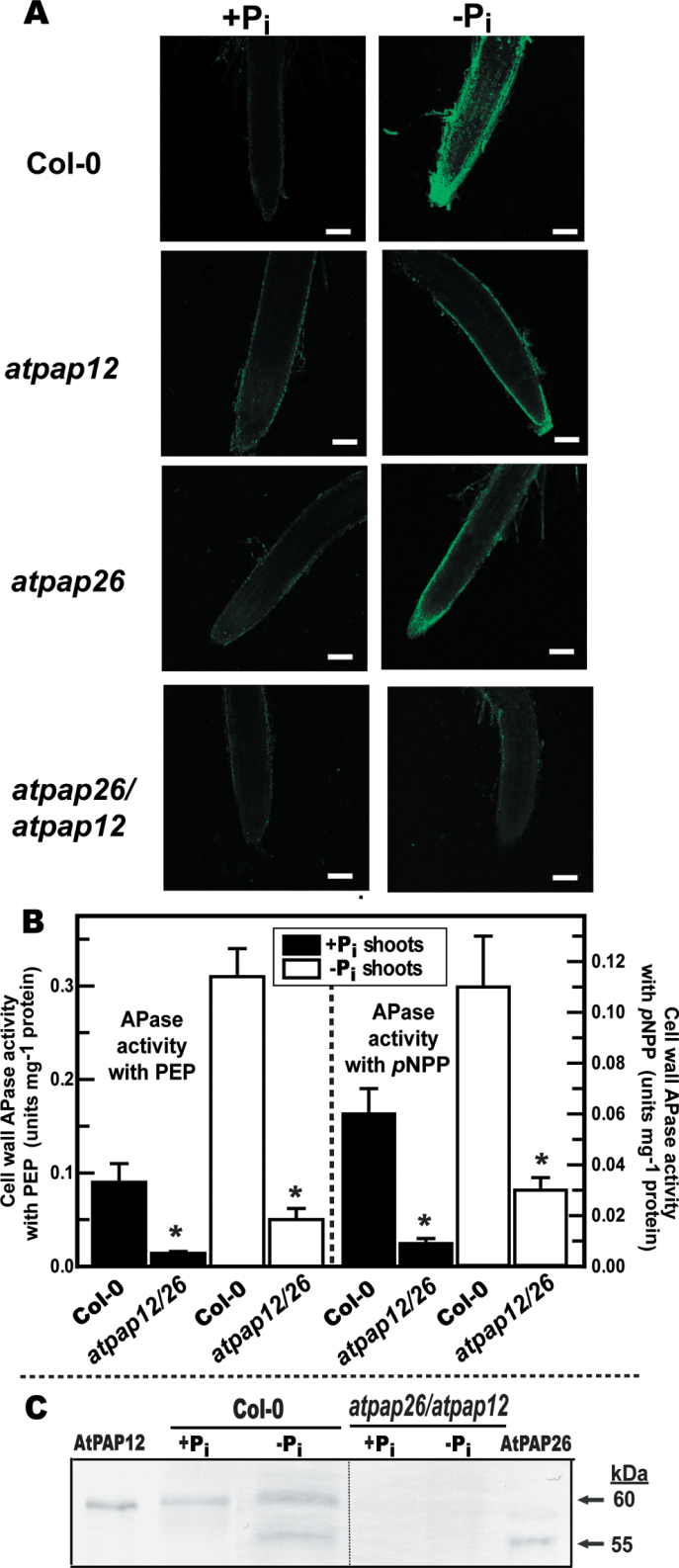
Fig. 3.
AtPAP12 and AtPAP26 make an important contribution to Pi-starvation-inducible APase activity of Arabidopsis root surfaces and shoot cell walls. (A) Histochemical staining of root-surface APase activity of Col-0, atpap12, atpap26 and atpap26/atpap12 seedlings using ELF-97 phosphate as a substrate. Green fluorescent precipitates of the APase product ELF-97 were observed using a confocal-laser scanning microscope. Bars, 100 µm. Seedlings were cultivated as described in the legend for Fig. 2B. (B) Concentrated cell-wall proteins extracted from shoots of Col-0 and atpap26/atpap12 seedlings were assayed for APase activity using 5mM PEP or 5mM pNPP as substrate. Values represent means ±SE of duplicate determinations on three biological replicates; asterisks indicate values that are significantly different from those of Col-0 (P <0.01). Seedlings were cultivated as described in the legend for Fig. 1. (C) Concentrated shoot cell-wall proteins (15 µg per lane) and purified native AtPAP26 and AtPAP12 (25ng per lane) (Tran et al., 2010a ) were subjected to immunoblot analysis with anti-AtPAP12 immune serum.