Abstract
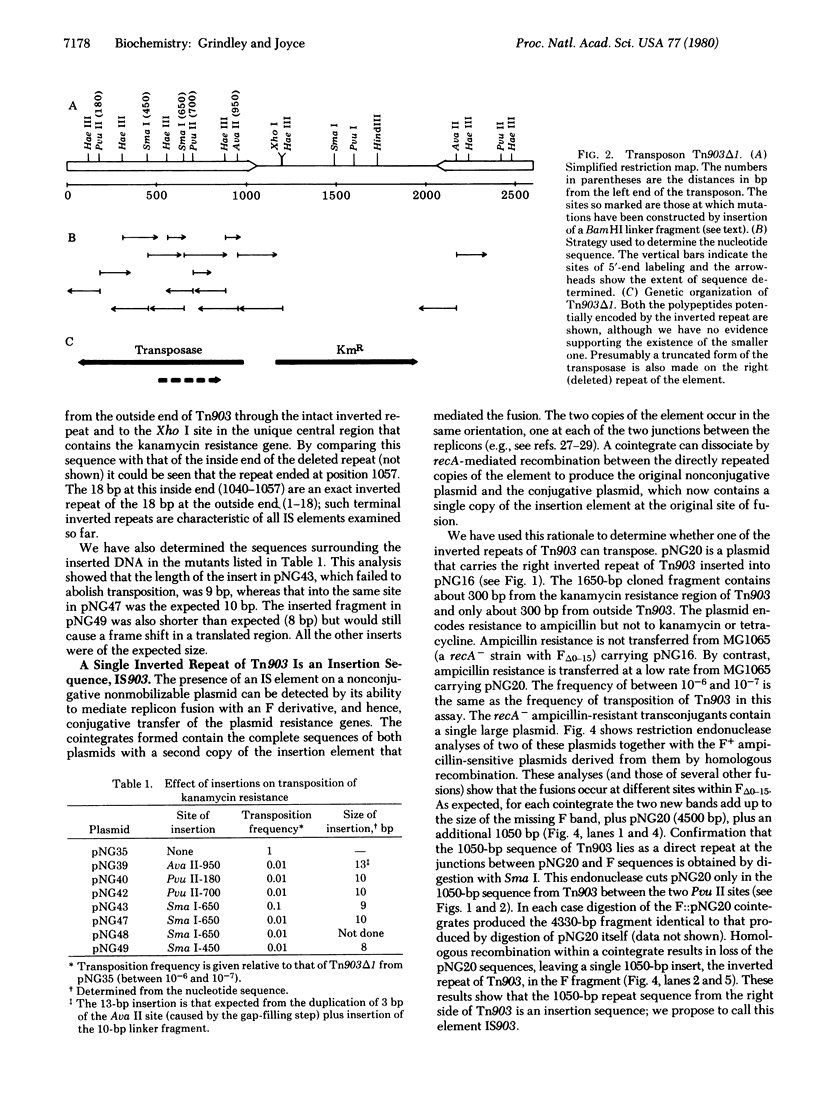
The kanamycin resistance transposon Tn903 consists of a unique region of about 1000 base pairs bounded by a pair of 1050-base-pair inverted repeat sequences. Each repeat contains two Pvu II endonuclease cleavage sites separated by 520 base pairs. We have constructed derivatives of Tn903 in which this 520-base-pair fragment is deleted from one or both repeats. Those derivatives that lack both 520-base-pair fragments cannot transpose, whereas those that lack just one remain transposition proficient. One such transposable derivative, Tn903 delta I, has been selected for further study. We have determined the sequence of the intact inverted repeat. The 18 base pairs at each end are identical and inverted relative to one another, a structure characteristic of insertion sequences. Additional experiments indicate that a single inverted repeat from Tn903 can, in fact, transpose; we propose that this element be called IS903. To correlate the DNA sequence with genetic activities, we have created mutations by inserting a 10-base-pair DNA fragment at several sites within the intact repeat of Tn903 delta 1, and we have examined the effect of such insertions on transposability. The results suggest that IS903 encodes a 307-amino-acid polypeptide (a "transposase") that is absolutely required for transposition of IS903 or Tn903.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrés I., Slocombe P. M., Cabello F., Timmis J. K., Lurz R., Burkardt H. J., Timmis K. N. Plasmid replication functions. II. Cloning analysis of the repA replication region of antibiotic resistance plasmid R6-5. Mol Gen Genet. 1979 Jan 5;168(1):1–25. doi: 10.1007/BF00267929. [DOI] [PubMed] [Google Scholar]
- Armstrong K. A., Hershfield V., Helinski D. R. Gene cloning and containment properties of plasmid Col E1 and its derivatives. Science. 1977 Apr 8;196(4286):172–174. doi: 10.1126/science.322277. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Childs G. J., Ohtsubo H., Ohtsubo E., Sonnenberg F., Freundlich M. Restriction endonuclease mapping of the Escherichia coli K12 chromosome in the vicinity of the ilv genes. J Mol Biol. 1977 Nov 25;117(1):175–193. doi: 10.1016/0022-2836(77)90030-4. [DOI] [PubMed] [Google Scholar]
- Chou J., Casadaban M. J., Lemaux P. G., Cohen S. N. Identification and characterization of a self-regulated repressor of translocation of the Tn3 element. Proc Natl Acad Sci U S A. 1979 Aug;76(8):4020–4024. doi: 10.1073/pnas.76.8.4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J., Lemaux P. G., Casadaban M. J., Cohen S. N. Transposition protein of Tn3: identification and characterisation of an essential repressor-controlled gene product. Nature. 1979 Dec 20;282(5741):801–806. doi: 10.1038/282801a0. [DOI] [PubMed] [Google Scholar]
- Clarke C. M., Hartley B. S. Purification, properties and specificity of the restriction endonuclease from Bacillus stearothermophilus. Biochem J. 1979 Jan 1;177(1):49–62. doi: 10.1042/bj1770049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Casadaban M. J., Chou J., Tu C. P. Studies of the specificity and control of transposition of the Tn3 element. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1247–1255. doi: 10.1101/sqb.1979.043.01.141. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan G., Saul M., Twigg A., Gill R., Sherratt D. Polypeptides expressed in Escherichia coli K-12 minicells by transposition elements Tn1 and Tn3. J Bacteriol. 1979 Apr;138(1):48–54. doi: 10.1128/jb.138.1.48-54.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faelen M., Toussaint A., De Lafonteyne J. Model for the enchancement of lambde-gal integration into partially induced Mu-1 lysogens. J Bacteriol. 1975 Mar;121(3):873–882. doi: 10.1128/jb.121.3.873-882.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill R. E., Heffron F., Falkow S. Identification of the protein encoded by the transposable element Tn3 which is required for its transposition. Nature. 1979 Dec 20;282(5741):797–801. doi: 10.1038/282797a0. [DOI] [PubMed] [Google Scholar]
- Gill R., Heffron F., Dougan G., Falkow S. Analysis of sequences transposed by complementation of two classes of transposition-deficient mutants of Tn3. J Bacteriol. 1978 Nov;136(2):742–756. doi: 10.1128/jb.136.2.742-756.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer M. S. The gamma delta sequence of F is an insertion sequence. J Mol Biol. 1978 Dec 15;126(3):347–365. doi: 10.1016/0022-2836(78)90045-1. [DOI] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Heffron F., So M., McCarthy B. J. In vitro mutagenesis of a circular DNA molecule by using synthetic restriction sites. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6012–6016. doi: 10.1073/pnas.75.12.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershfield V., Boyer H. W., Chow L., Helinski D. R. Characterization of a mini-ColC1 plasmid. J Bacteriol. 1976 Apr;126(1):447–453. doi: 10.1128/jb.126.1.447-453.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nomura N., Yamagishi H., Oka A. Isolation and characterization of transducing coliphage fd carrying a kanamycin resistance gene. Gene. 1978 Feb;3(1):39–51. doi: 10.1016/0378-1119(78)90006-9. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ohmori H., Ohtsubo E. Nucleotide-sequence analysis of Tn3 (ap): implications for insertion and deletion. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1269–1277. doi: 10.1101/sqb.1979.043.01.144. [DOI] [PubMed] [Google Scholar]
- Oka A., Nomura N., Sugimoto, Sugisaki H., Takanami M. Nucleotide sequence at the insertion sites of a kanamycin transposon. Nature. 1978 Dec 21;276(5690):845–847. doi: 10.1038/276845a0. [DOI] [PubMed] [Google Scholar]
- Parker R. C., Watson R. M., Vinograd J. Mapping of closed circular DNAs by cleavage with restriction endonucleases and calibration by agarose gel electrophoresis. Proc Natl Acad Sci U S A. 1977 Mar;74(3):851–855. doi: 10.1073/pnas.74.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Rothstein S. J., Jorgensen R. A., Postle K., Reznikoff W. S. The inverted repeats of Tn5 are functionally different. Cell. 1980 Mar;19(3):795–805. doi: 10.1016/s0092-8674(80)80055-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A., MacHattie L. A. Integration and excision of prophage lambda mediated by the IS 1 element. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1135–1142. doi: 10.1101/sqb.1979.043.01.127. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Cohen S. N., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973 Apr 5;75(2):235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Hsu M. T., Otsubo E., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. I. Structure of F-prime factors. J Mol Biol. 1972 Nov 14;71(2):471–497. doi: 10.1016/0022-2836(72)90363-4. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So M., Heffron F., McCarthy B. J. The E. coli gene encoding heat stable toxin is a bacterial transposon flanked by inverted repeats of IS1. Nature. 1979 Feb 8;277(5696):453–456. doi: 10.1038/277453a0. [DOI] [PubMed] [Google Scholar]
- Staden R. Sequence data handling by computer. Nucleic Acids Res. 1977 Nov;4(11):4037–4051. doi: 10.1093/nar/4.11.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starlinger P. IS elements and transposons. Plasmid. 1980 May;3(3):241–259. doi: 10.1016/0147-619x(80)90039-6. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Takeya T., Nomiyama H., Miyoshi J., Shimada K., Takagi Y. DNA sequences of the integration sites and inverted repeated structure of transposon Tn3. Nucleic Acids Res. 1979;6(5):1831–1841. doi: 10.1093/nar/6.5.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]