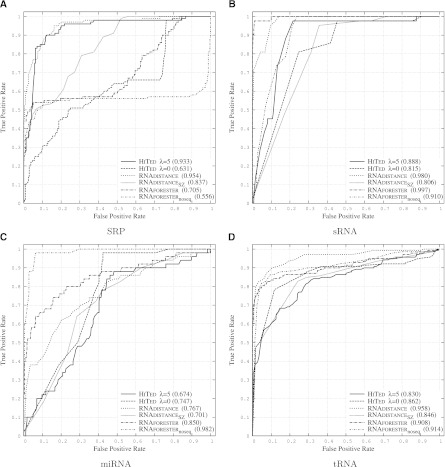
FIGURE 4.
ROC plots comparing HiTed, RNAdistance, and RNAforester on data sets SRP (A), sRNA (B), miRNA (C), and tRNA (D). The noise data used in all four cases are random genome segments from viral genomes. RNAdistance was used with default parameters (tree edit distance on full structure representation) and with coarse-grained tree edit distance (Shapiro and Zhang 1990) RNAdistanceSZ. RNAforester was used with default parameters and without scoring sequence similarity (RNAforesternoseq). HiTed was used with the indicated λ-values.