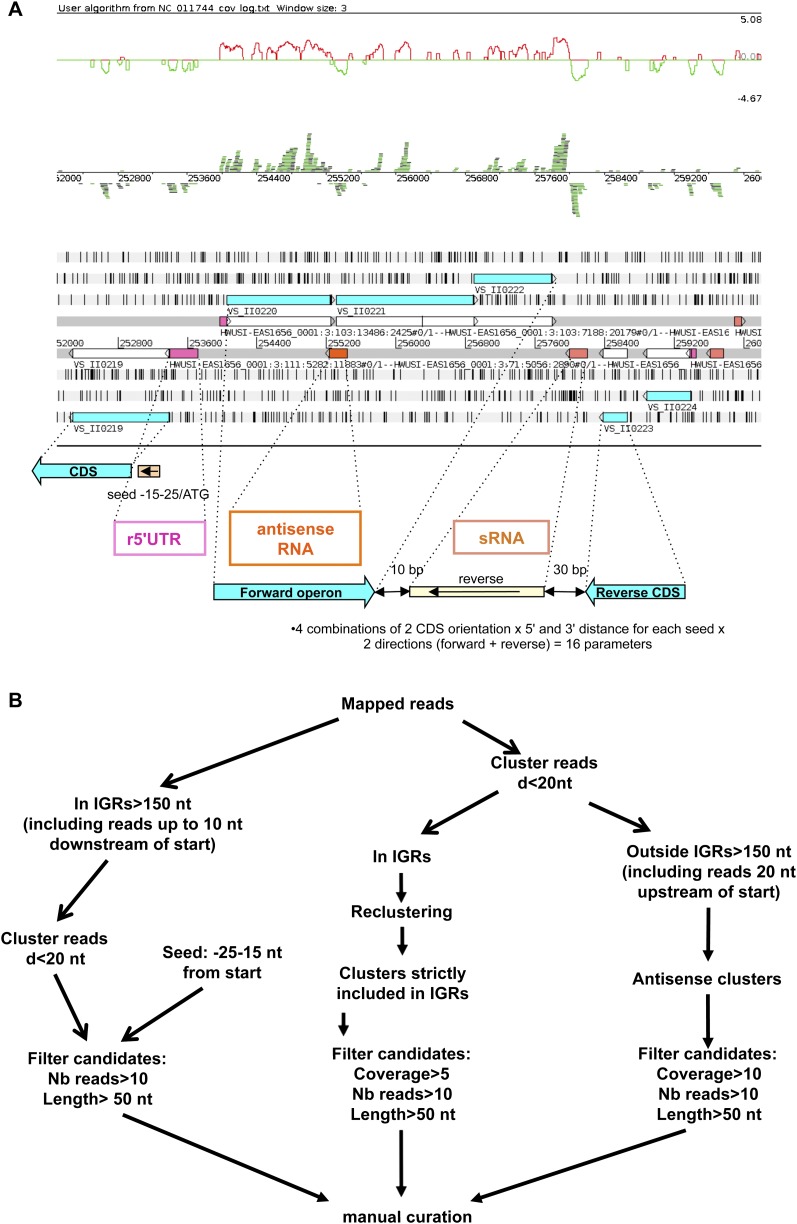
FIGURE 2.
Visualization interface and models for the three kinds of sRNA candidates. (A) We used the Artemis sequence editor tool to check the regulatory RNA candidates manually. We added two tracks to the Artemis classical view of genomic information: the ln of read coverage track (top) and the BamView of the mapped reads (middle). Forward sequences and reverse sequences are above and under the line, respectively. (White boxes) CDSs; (blue boxes) annotated proteins; (pink boxes) cis-encoded r5′UTRs; (orange boxes) cis-encoded asRNAs; (salmon boxes) trans-encoded sRNAs. Models for parameter definition are diagrammatically outlined for each category of regulatory RNAs under the Artemis window. See Materials and Methods and Results sections for details. (B) Schematic representation of the different steps in script execution. r5′UTR, sRNA, and asRNA computational workflows are depicted, respectively, by the left, middle, and right pathways. Most of the steps are managed using one or more S-MART tools. (Nb reads) Number of reads; (d) distance below which two clusters of overlapping reads are merged.