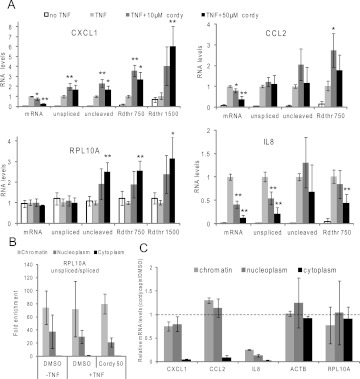
FIGURE 6.
Effects of cordycepin on nascent and mature mRNA. (A) RNA was isolated from ASM cells. RT-qPCR was performed for mature mRNA (mRNA), unspliced pre-mRNA (unspliced), uncleaved pre-mRNA (uncleaved), and for runthrough RNA transcribed from the terminator region, 750 or 1500 nt from the cleavage site (Rdthr750 and Rdthr1500). The location of the primers used is indicated in Figure 5B. mRNA levels were normalized to GAPDH mRNA levels. All error bars in this figure are standard deviations over three biological replicates. Statistical significance in a paired T-test for comparison with noncordycepin treated is indicated: (*) P ≤ 0.05, (**) P ≤ 0.01. (B) Chromatin associated, nucleoplasmic and cytoplasmic fractions are successfully separated. RNA fractions were isolated and RT-qPCR for spliced and unspliced RPL10A mRNA was performed. The ratio of unspliced over spliced for each sample was divided by this ratio in the cytoplasmic fraction. Error bars represent standard deviations over three biological replicates. (C) The effect of cordycepin (Cordy) on inflammatory and control mRNA levels in chromatin-associated, nucleoplasmic, and cytoplasmic RNA isolated from TNF-treated ASM cells, as determined by RT-qPCR. Data were normalized to GAPDH mRNA levels, and the ratio between cordycepin-treated and noncordycepin-treated samples was graphed (a value of 1 therefore indicating no change). Error bars represent standard deviations over three biological replicates.