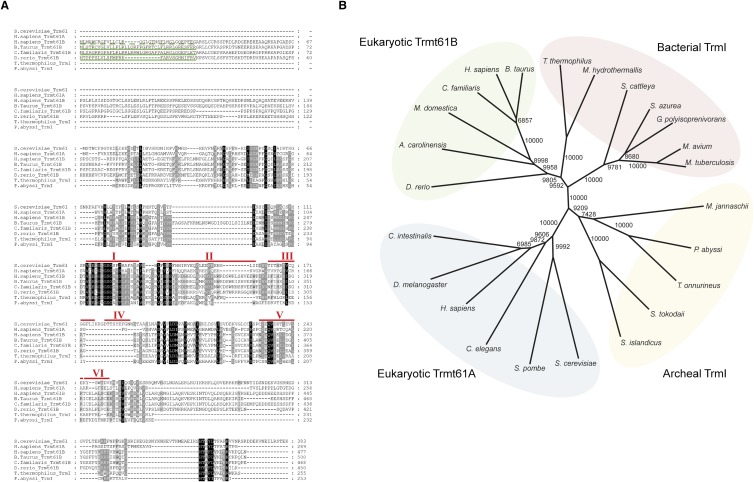
FIGURE 6.
Sequence alignment and phylogenetic analysis of Trmt61B and TrmI family proteins. (A) Sequence alignment of Trmt61B and its homologs from Eukarya, Bacteria, and Archaea. RefSeq or UniProt IDs of these proteins are listed in Supplemental Table S2. The mitochondria-targeting signal (MTS) was predicted using Mitoprot software (Claros and Vincens 1996) and is shown by green underlines. Human MTS is depicted as a dashed line, because the exact end of the MTS could not be predicted. Motifs I–VI of the Rossmann-fold-type methyltransferase in S. cerevisiae Trm61 (Kozbial and Mushegian 2005) are indicated. (B) A nonrooted neighbor-joining tree for Trmt61B and TrmI family proteins was constructed by Clustal_X software (Thompson et al. 1997). The bootstrap value was obtained by 10,000 trials and is indicated at each node. RefSeq or UniProt IDs are listed in Supplemental Table S2.