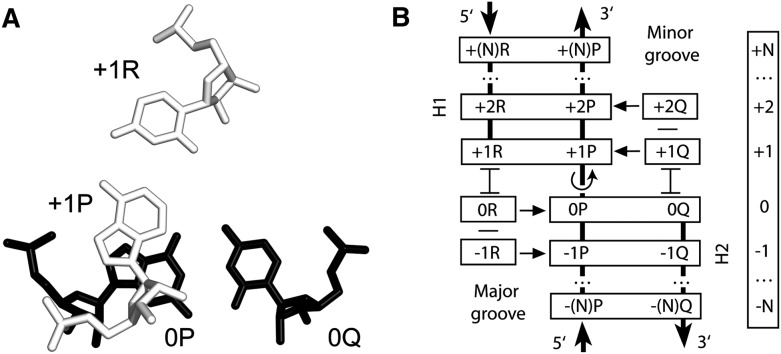
FIGURE 1.
Definition of a TJ arrangement. (A) A typical juxtaposition of 2 bp, [0P;0Q] (black) and [+1P;+1R] (white), forming a TJ. Nucleotide +1P is next to 0P in the polynucleotide chain, while nucleotides 0Q and +1R are not directly connected. Compared to the standard A-RNA conformation, there is an over-twist between 0P and +1P (in the figure, it is ∼35°), which reduces the area of stacking between the 2 nt. (B) Schematic representation of a TJ arrangement. All individual nucleotides (short horizontally oriented rectangles) and base pairs (long rectangles) are positioned in layers. Each layer has its own number, as indicated in the vertical bar on the right. Base pairs [+1R;+1P] and [+2R;+2P] form helix 1 (H1), while base pairs [0P;0Q] and [−1P;−1Q] form helix 2 (H2). The over-twist occurs between base pairs [+1R;+1P] and [0P;0Q], as indicated by the rotating arrow. Unpaired nucleotides +1Q and +2Q stack on 0Q and interact with the minor groove of H1 (indicated by horizontal arrows). Nucleotides 0R and −1R stack on +1R and interact with the major groove of H2. Nucleotides 0R and +1Q make the direct stabilization of the over-twist. Outside double helical regions, the interbase stacking is indicated either by letter “H” rotated for 90° (symbol  ) or by a short horizontal line.
) or by a short horizontal line.