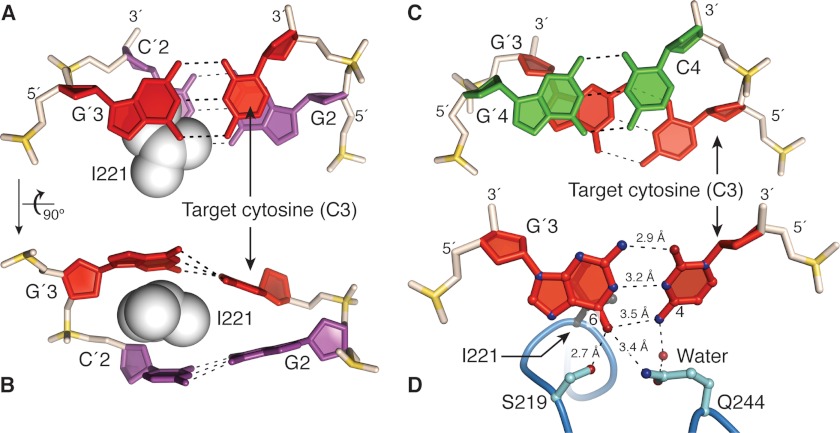
FIGURE 4.
M.HaeIII-induced destabilization of the target cytosine, C3. A and B, two different close-up views of the target base pair C3/G′3 and its unstacked neighbor G2/C′2. The side chain of the intercalating residue Ile-221 is shown in space-filling representation. Note that the target cytosine C3 is partially destacked from G2, whereas the complementary nucleobases G′3 and C′2 are completely destacked through Ile-221 intercalation. Note also the distorted base pairing of the target base pair. C, close-up view of stacking between base pairs 3 and 4. Note that the two migratory cytosines, C3 and C4 are completely destacked by under-rotation of the DNA helix at that base pair step. D, direct contacts in InC to the target base pair. The interaction between G′3 and Ser-219 in InC is maintained in ExC. Gln-244 is well positioned to compete for the hydrogen bonding of the target cytosine C3 with the complementary guanine G′3. The water-mediated interaction of Gln-244 with C3 in InC, which has poor geometry, is replaced in ExC by a direct and geometrically favorable contact to the frameshifted nucleobase, C4 (data not shown). The orientation in D is slightly different from that on A for clarity.