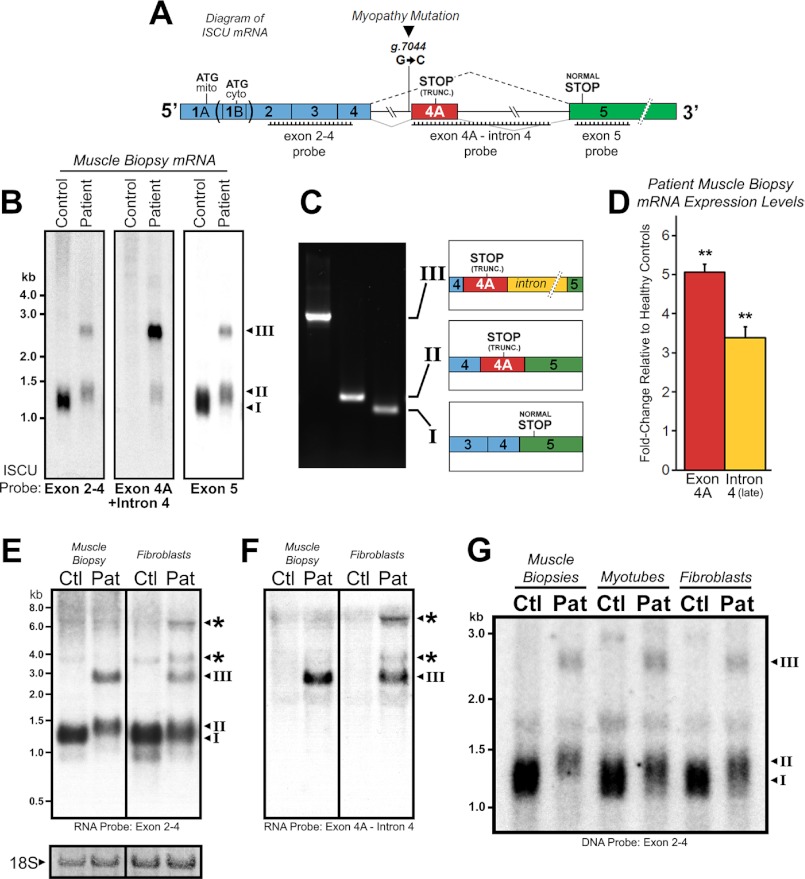
FIGURE 1.
Altered ISCU mRNA expression patterns in ISCU myopathy patients. A, a schematic representation of ISCU mRNA showing the single intronic point mutation within the fourth intron, canonical, and aberrant splicing patterns and regions of Northern blot probe complementarity. mito, mitochondrial; cyto, cytosolic; TRUNC., truncated. B, hybridization of ISCU-specific 32P-labeled DNA probes with control and patient vastus lateralis biopsy RNA demonstrated normal (I) and additional patient-specific (II and III) ISCU mRNA bands. C, RT-PCR and sequencing using primers flanking ISCU exons 4 and 5 confirmed the identity of the three major ISCU bands (I–III), with transcript II containing 100 bp and transcript III containing 1173 bp of intronic sequence. D, qRT-PCR analysis of three ISCU myopathy patients and seven control biopsies using primer sets for ISCU exon 4A and downstream intron 4 sequences. E and F, Northern blot analysis of ISCU mRNA expression in control (Ctl) and patient (Pat) muscle biopsies and fibroblasts using high specific activity RNA probes revealed several larger ISCU-specific mRNA bands in ISCU myopathy patient fibroblasts. Methylene blue-stained 18 S ribosomal RNA served as a loading control. G, Northern blot analysis of control and patient muscle biopsies alongside myotubes and fibroblasts demonstrates a decreased residual level of normally spliced ISCU mRNA in the patient muscle biopsy.