Background: Phosphatidylethanolamine is proposed to regulate mitochondrial fusion, but its mechanism of action is unknown.
Results: Decreasing phosphatidylethanolamine reduces the rate of lipid mixing and the biogenesis of Mgm1, a mitochondrial fusion protein.
Conclusion: Psd1 regulates the lipid and protein machineries of mitochondrial fusion.
Significance: Understanding how lipid metabolism regulates mitochondrial dynamics will reveal its role in cellular functions such as apoptosis and autophagy.
Keywords: Lipid Metabolism, Membrane Fusion, Mitochondria, Phosphatidylethanolamine, Protein Processing
Abstract
Non–bilayer-forming lipids such as cardiolipin, phosphatidic acid, and phosphatidylethanolamine (PE) are proposed to generate negative membrane curvature, promoting membrane fusion. However, the mechanism by which lipids regulate mitochondrial fusion remains poorly understood. Here, we show that mitochondrial-localized Psd1, the key yeast enzyme that synthesizes PE, is required for proper mitochondrial morphology and fusion. Yeast cells lacking Psd1 exhibit fragmented and aggregated mitochondria with impaired mitochondrial fusion during mating. More importantly, we demonstrate that a reduction in PE reduces the rate of lipid mixing during fusion of liposomes with lipid compositions reflecting the mitochondrial membrane. This suggests that the mitochondrial fusion defect in the Δpsd1 strain could be due to the altered biophysical properties of the mitochondrial membrane, resulting in reduced fusion kinetics. The Δpsd1 strain also has impaired mitochondrial activity such as oxidative phosphorylation and reduced mitochondrial ATP levels which are due to a reduction in mitochondrial PE. The loss of Psd1 also impairs the biogenesis of s-Mgm1, a protein essential for mitochondrial fusion, further exacerbating the mitochondrial fusion defect of the Δpsd1 strain. Increasing s-Mgm1 levels in Δpsd1 cells markedly reduced mitochondrial aggregation. Our results demonstrate that mitochondrial PE regulates mitochondrial fusion by regulating the biophysical properties of the mitochondrial membrane and by enhancing the biogenesis of s-Mgm1. While several proteins are required to orchestrate the intricate process of membrane fusion, we propose that specific phospholipids of the mitochondrial membrane promote fusion by enhancing lipid mixing kinetics and by regulating the action of profusion proteins.
Introduction
Mitochondria are highly dynamic organelles, constantly undergoing fusion and fission reactions to maintain a tubular network. Whereas the protein machineries of mitochondrial fusion and fission have been extensively studied, the role of lipids in regulating these processes is just beginning to be uncovered. It has been shown that non–bilayer-forming lipids such as phosphatidic acid, cardiolipin (CL),3 and phosphatidylethanolamine (PE) are required for proper mitochondrial morphology (1–6). Recently, a study examining the loss of both CL and PE by deleting CL synthase, CRD1, and the mitochondrial phosphatidylserine decarboxylase, PSD1, respectively, demonstrated that both lipids are required for mitochondrial morphology and fusion (5). In a mouse model, disrupting the mammalian homolog of PSD1, Pisd, resulted in abnormal mitochondrial morphology (6). Furthermore, a reduction in the levels of a mitochondrial-localized phospholipase D (mitoPLD, which synthesizes phosphatidic acid from the hydrolysis of CL) resulted in mitochondrial fragmentation (1). It is proposed that these non–bilayer-forming lipids induce hexagonal phases, generating negative membrane curvature that promotes membrane fusion (7, 8). However, the mechanisms by which these lipids regulate mitochondrial dynamics remain poorly understood.
The protein machinery of mitochondrial fusion is highly conserved from yeast to mammals and consists of large dynamin-like GTPases. In yeast, the outer mitochondrial membrane (OMM)-localized Fzo1 (Mfn1/2 in mammals) mediates OMM fusion, whereas the inner mitochondrial membrane (IMM)-localized Mgm1 (OPA1 in mammals) mediates IMM fusion (9–12). A fungal-specific protein, Ugo1, serves as an adaptor, tethering Fzo1 and Mgm1, coordinating OMM and IMM fusion (13). Mgm1 exists as two isoforms, long (l-Mgm1) and short Mgm1 (s-Mgm1) (14). A proper balance of l- and s-Mgm1 protein levels is crucial for mitochondrial fusion; perturbing this ratio results in impaired mitochondrial fusion (15). The formation of s-Mgm1 from full-length Mgm1 (FL-Mgm1) requires the enzymatic activity of the mitochondrial rhomboid Rbd1/Pcp1 and is an ATP-dependent process (14, 16, 17). Here, we show that Psd1 is required for maintaining mitochondrial ATP levels and for the biogenesis of s-Mgm1 but does not impinge on Rbd1 activity. We also provide evidence that Psd1 regulates mitochondrial fusion by regulating the biophysical properties of the mitochondrial membrane, likely enhancing the rate of lipid mixing during fusion. Together, our findings reveal the mechanisms by which Psd1-synthesized PE promotes mitochondrial fusion and demonstrate a complex interaction between lipid homeostasis, mitochondrial dynamics, and mitochondrial activity.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, and Media
All strains used in this study are derivatives of BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0). Gene deletions and genomic tagging were performed by homologous recombination of gene-specific PCR products. pYX122-mtGFP (mtGFP), pYES-mtGFP and pYES-mtBFP (galactose-inducible mtGFP and mtBFP, respectively) were kind gifts from Dr. Benedikt Westermann and have been described previously (18). pRS315-s*Mgm1–3×HA (s*Mgm1) was generated by cloning a SacI/SalI double-digested fragment from pRS314-s*Mgm1–3×HA (a gift from Dr. Andreas Reichert and has been described previously (15)) into pRS315. pRS315-Mgm1-G100D was also a gift from Dr. Andreas Reichert and was described previously (17). Synthetic minimal medium consisted of 0.17% yeast nitrogen base, 5% ammonium sulfate, and 2% glucose, galactose or glycerol as the carbon source. Rich medium consisted of 1% yeast extract, 2% peptone, and 2% glucose or galactose. Ethanolamine supplementation was added to a final concentration of 5 mm.
In Vivo Mitochondrial Fusion Assay
MATa cells expressing pYES-mtGFP and MATα cells expressing pYES-mtBFP were grown to log phase in synthetic galactose medium to induce the expression of mtGFP and mtBFP. An equal number of cells were mixed together and washed twice with synthetic glucose medium to stop mtGFP and mtBFP production. Cells were resuspended in the same volume of synthetic glucose medium, mixed thoroughly, and spread evenly onto a synthetic glucose-agar plate using a sterile inoculating loop. Liquid medium on the agar plate was air-dried, and the plate was incubated for 3 h at 30 °C for yeast mating. Cells were then carefully scraped off the plate with a sterile inoculating loop, resuspended in synthetic glucose medium, and analyzed by fluorescence microscopy.
In Vitro Liposome Fusion Assay
Phospholipid compositions of WT and Δpsd1 mitochondria were determined previously (19). For fluorescence-labeled liposomes, 1.6% of the PE was replaced with 0.8% NBD-PE and 0.8% Lissamine Rhodamine B-PE (Avanti Polar Lipids). Lipids were mixed in a glass tube and dried under a gentle stream of nitrogen. Residual chloroform was removed under a rotary evaporator for 90 min. Lipid films were rehydrated to a concentration of 0.5 mm in liposome buffer (20 mm HEPES, pH 7.5, 150 mm NaCl) at 65 °C for 1 h, vortexing every 15 min for homogeneity. To make liposomes, hydrated lipids were extruded 15 times through a 1.0-μm filter membrane (Avanti Polar Lipids). Liposome fusion was carried out in 96-well black plates in 100-μl reactions. 1 μl of labeled liposomes was mixed with 9 μl of unlabeled liposomes in 80 μl of liposome buffer. After reading basal fluorescence for 5 min, 10 μl of 1.0 m CaCl2 was added to induce liposome fusion. Liposome buffer was used as a negative control, and maximum lipid mixing was determined by adding 10 μl of 10% octaethylene glycol monododecyl ether (C12E8). Fluorescence was normalized to the initial fluorescence reading and presented as a percentage of maximum lipid mixing.
Oxidative Phosphorylation
Cells were grown to log phase in rich galactose medium. 3 × 107 cells were harvested per reading, pelleted, and resuspended in 1 ml of fresh fully oxygenated medium for oxygen consumption measurements using a Clark-type oxygen electrode (Strathkelvin Instruments). 100% ethanol was added to a final concentration of 1% (v/v). After the rate of oxygen consumption stabilized, 2 mm carbonylcyanide m-chlorophenylhydrazone was added to a final concentration of 8 μm to induce maximum oxygen consumption.
Mitochondrial Purification
Cells were grown to log phase in 50 ml of rich galactose medium, and mitochondria were purified as described by Gregg et al. with slight modifications (20). Briefly, after mitochondrial enrichment, pellets were resuspended in 1 ml of resuspension buffer (homogenization buffer without BSA) and gently homogenized three times with a tight Dounce. Suspensions were then pelleted at 3,000 × g for 5 min at 4 °C. The resulting supernatants were then centrifuged at 12,000 × g for 15 min at 4 °C to pellet mitochondria. Mitochondrial purity was assessed by Western blotting.
ATP Assay
Purified mitochondria were resuspended in 15 μl of 5.5% TCA, 2 mm EDTA and incubated on ice for 10 min to extract ATP and precipitate mitochondrial proteins. Extracted ATP was separated from precipitated proteins by centrifugation at 16,100 × g for 10 min at 4 °C. Supernatants containing ATP were collected for ATP measurements using an ATP Determination kit (Invitrogen) according to the manufacturer's instructions. Precipitated proteins were washed twice with ice-cold acetone and heated at 95 °C for 5 min to fully evaporate the acetone. Pellets were then resuspended in 15 μl of reducing sample buffer and boiled for 5 min at 95 °C. Protein concentration was determined using the RC DC protein assay (Bio-Rad).
Cycloheximide Chase
Yeast cells were grown to log phase in rich galactose medium. Cycloheximide was added to a final concentration of 100 μg/ml. Cells were collected at the indicated time points and lysed by alkaline lysis for Western blot analysis. Lysates from 5 × 106 cells were loaded per sample. Proteins were detected using Mgm1, Fzo1, and Tom40 antisera, gifts from Dr. Jodi Nunnari, Dr. Andreas Reichert, and Dr. Thomas Langer, respectively.
RESULTS
Psd1 Is Required for Normal Mitochondrial Morphology
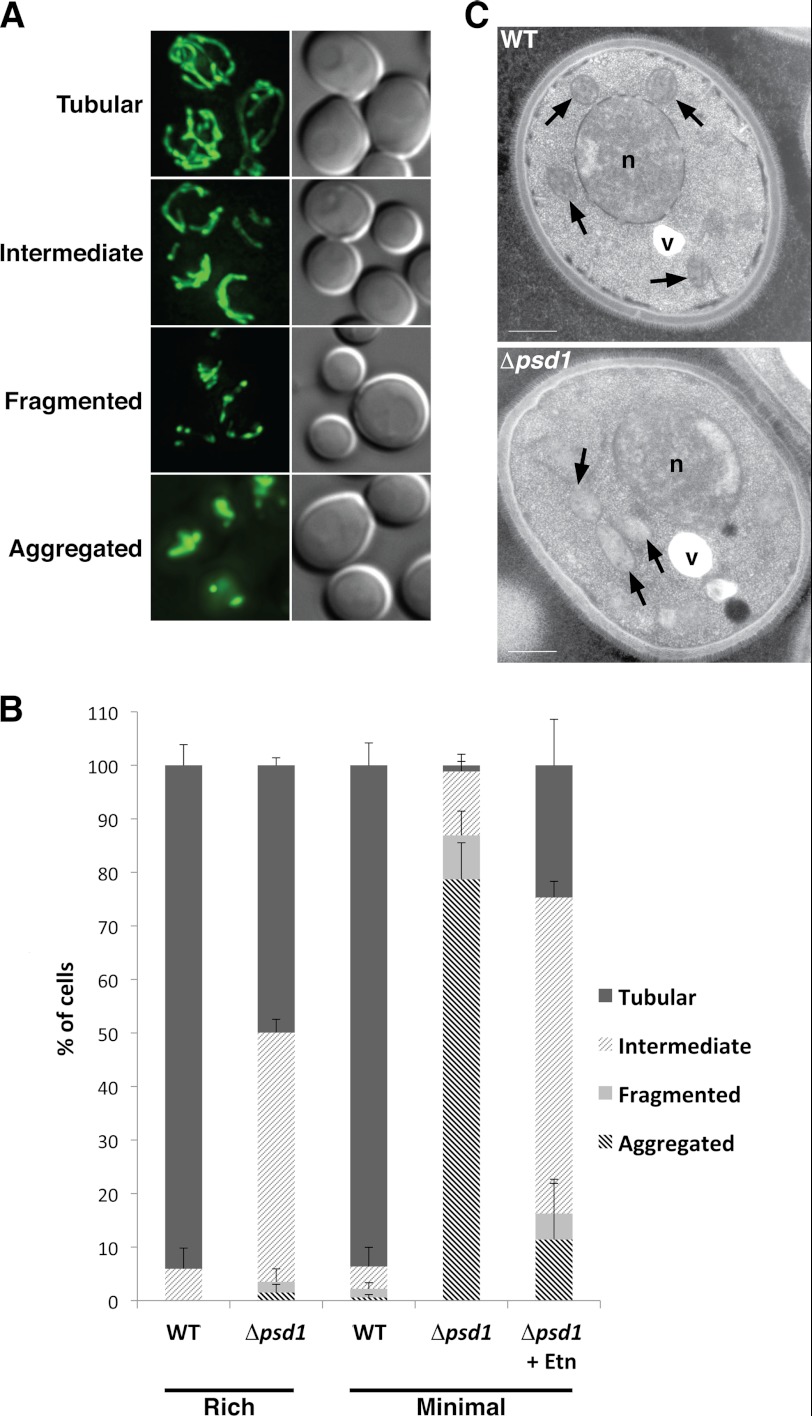
To determine whether phospholipid composition plays a role in regulating mitochondrial membrane dynamics, we examined the mitochondrial morphology of the Δpsd1 yeast strain. A previous study demonstrated that the loss of Psd1 results in an alteration in the mitochondrial phospholipid composition when cells were cultured in rich medium with lactate as a carbon source. Despite the altered mitochondrial lipid composition, it was reported that the mitochondrial morphology of these cells was unaffected (21). Indeed, we found that Δpsd1 cells cultured in rich medium had mitochondria that are tubular, somewhat similar to that of wild type (WT) (Fig. 1, A and B). However, upon further characterization, we find that 50% of Δpsd1 cells had an intermediate mitochondrial morphology (Fig. 1, A and B) consisting of tubules, but lacking an organized interconnected network that is seen in WT yeast cells. Because rich medium contains additional nutrients that could affect overall lipid metabolism, we cultured cells in minimal medium, supplying only the nutrients necessary for survival. In contrast to cells cultured in rich medium, 85% of Δpsd1 cells cultured in minimal medium had mitochondria that were fragmented and aggregated (Fig. 1, A and B). This result is consistent with recent findings that Psd1 is required for normal mitochondrial morphology (4, 5). To further define the mitochondrial aggregation phenotype, we performed electron microscopic analysis. Unlike WT mitochondria that are evenly distributed throughout the cytoplasm, Δpsd1 mitochondria were clustered but not fused (Fig. 1C). Furthermore, the inner membrane of Δpsd1 mitochondria was less electron-dense, suggesting possible inner membrane defects (Fig. 1C). Together, these data indicate that Psd1 is required for normal mitochondrial morphology, highlighting the importance of mitochondrial lipid metabolism in maintaining organellar morphology. The clustered mitochondrial phenotype of the Δpsd1 strain resembles that of mitochondrial fusion mutants (22), suggesting that Psd1 might regulate mitochondrial fusion.
FIGURE 1.
Psd1 is required for normal mitochondrial morphology. WT and Δpsd1 cells were transformed with a mitochondrial matrix-targeted GFP construct (mtGFP) and cultured in rich and minimal media with galactose as the carbon source. A, representative images of cells with tubular, intermediate, fragmented, and aggregated mitochondria. B, quantification of the mitochondrial morphology of cells cultured in rich and minimal media. More than 300 cells were analyzed per strain. Error bars represent the S.D. of three independent experiments. C, electron microscopic analysis of the WT and Δpsd1 strains cultured in minimal medium. Mitochondria (arrows), nuclei (n), and vacuoles (v) are indicated. Scale bars, 500 nm.
Psd1 Is Required for Proper Mitochondrial Fusion during Yeast Mating
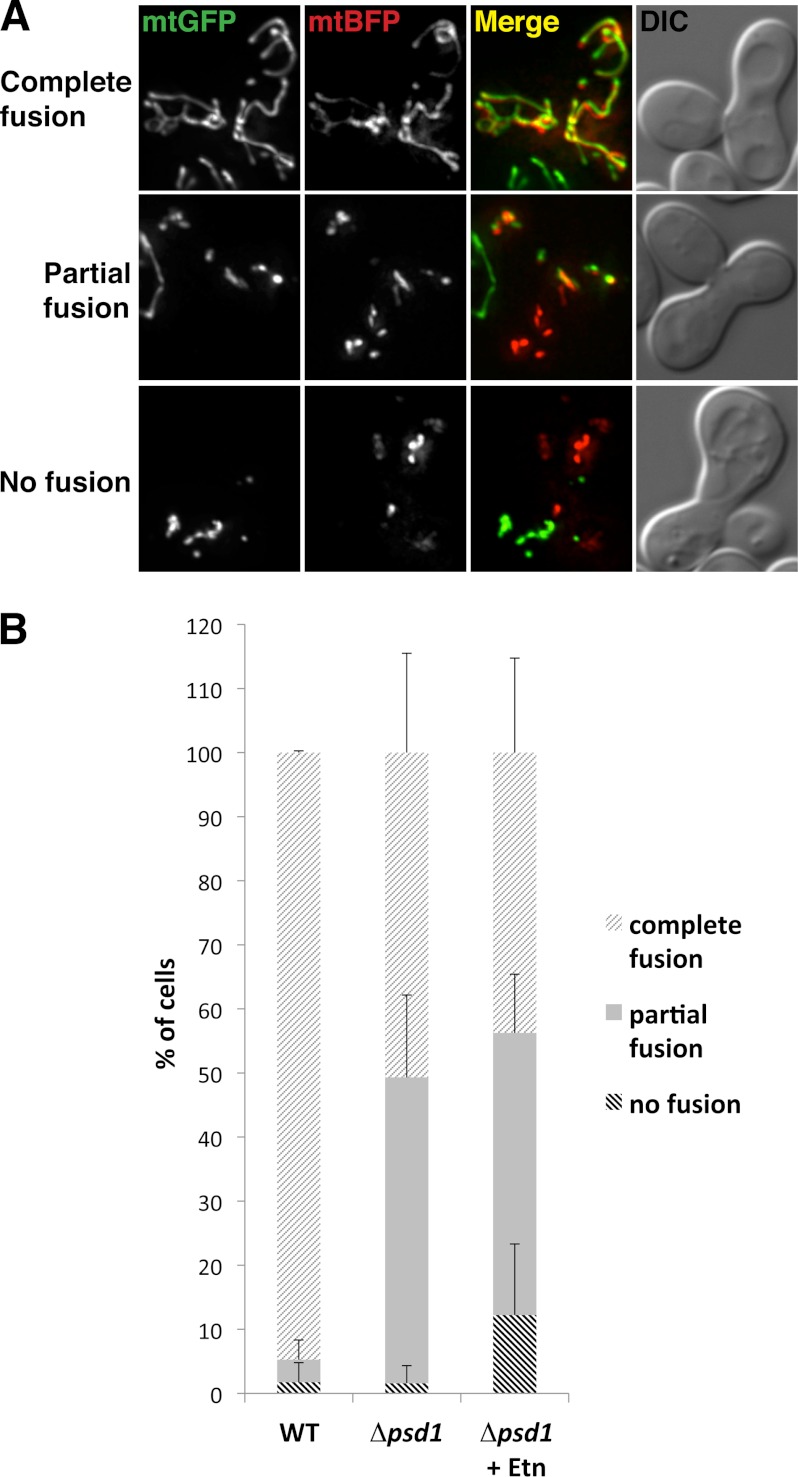
To assess whether the mitochondrial morphology defect in Δpsd1 cells is due to a defect in mitochondrial fusion, we performed an in vivo mitochondrial fusion assay. During yeast mating, mitochondria fuse, and mitochondrial content mixing of the mating MATa and MATα haploids can be used as an assay to monitor mitochondrial fusion. As a positive control, WT cells had complete mixing of mitochondrial content (Fig. 2, A and B), indicating complete mitochondrial fusion in this assay. In contrast, only 50% of Δpsd1 cells showed complete mitochondrial content mixing whereas the remaining 50% had only partial mixing (Fig. 2, A and B). This result is surprising considering the mitochondrial fragmentation and aggregation in the Δpsd1 strain (Fig. 1B). Nevertheless, the ability of Δpsd1 mitochondria to undergo fusion indicates that Psd1 is not an obligate member of the mitochondrial fusion machinery. However, the partial fusion phenotype strongly implicates Psd1 as an important regulator and that the altered phospholipid composition of the mitochondrial membrane affects in vivo mitochondrial fusion during mating.
FIGURE 2.
Psd1 is required for mitochondrial fusion during yeast mating. A, representative images of cells that have undergone complete, partial, and no mitochondrial fusion during yeast mating. Haploid WT and Δpsd1 strains were transformed with a galactose-inducible mtGFP or mtBFP. Strains were cultured and mated as described under “Experimental Procedures” and analyzed for mitochondrial content mixing. The extent of mitochondrial fusion was indicated by the amount of mixing between mtGFP and mtBFP. B, quantification of zygotes that have undergone complete, partial, or no mitochondrial fusion. 20 zygotes were analyzed per strain. Error bars, S.D.
Phospholipid Composition Affects the Rate of Lipid Mixing
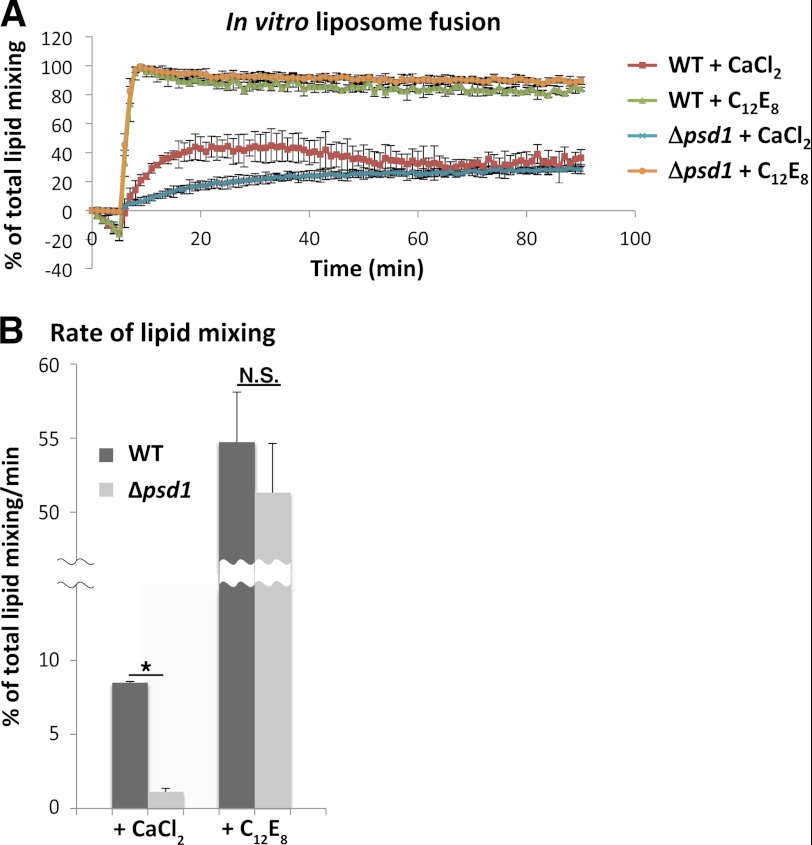
Impaired mitochondrial fusion of the Δpsd1 strain during yeast mating suggests that the phospholipid composition of the mitochondrial membrane plays an important role in promoting mitochondrial fusion. To assess whether the altered lipid composition affected the biophysical properties of the mitochondrial membrane that could influence mitochondrial fusion, we performed an in vitro liposome fusion assay using liposomes with phospholipid compositions similar to that of WT and Δpsd1 mitochondria (19). Upon the addition of CaCl2 to induce fusion, liposomes with phospholipid compositions similar to that of Δpsd1 mitochondria (Δpsd1 liposomes) fused to the same extent as those with compositions similar to that of WT mitochondria (WT liposomes). This indicates that altering the phospholipid composition does not affect the extent to which liposomes fused (Fig. 3A) and is consistent with our in vivo mitochondrial fusion assay that Δpsd1 mitochondria can undergo complete fusion (Fig. 2B). Interestingly, the slope of the curve during mixing of Δpsd1 liposomes is different from that of WT liposomes (Fig. 3A). Because the slopes of the curves represent the relative rates of reaction, we analyzed the slopes to determine whether reduced fusion kinetics could contribute to the mitochondrial fusion defect in the Δpsd1 strain. Our analysis indicates that Δpsd1 liposomes have an ∼75% reduction in their rates of lipid mixing compared with WT liposomes (Fig. 3B). Due to the absence of proteins in this assay, our result indicates that changes in the phospholipid content of Δpsd1 mitochondria likely alter the biophysical properties of the mitochondrial membrane, reducing the kinetics of lipid mixing during mitochondrial fusion.
FIGURE 3.
Liposomes with lipid compositions similar to Δpsd1 mitochondria have a reduced rate of lipid mixing. A, in vitro liposome fusion was performed as described under “Experimental Procedures.” Liposomes labeled with NBD and Lissamine Rhodamine B were mixed with unlabeled liposomes; basal NBD fluorescence was monitored for 5 min. CaCl2 and C12E8 were added to induce fusion and maximum lipid mixing, respectively. The increase in NBD fluorescence indicates relief of NDB quenching by Lissamine Rhodamine B. B, quantification of the rates of lipid mixing indicated by the slopes of the curves in A. Values represent the mean ± S.D. (error bars) of three independent experiments. *, p < 0.0001; N.S., not significant (p > 0.05).
Psd1 Is Required for Proper Mitochondrial Activity
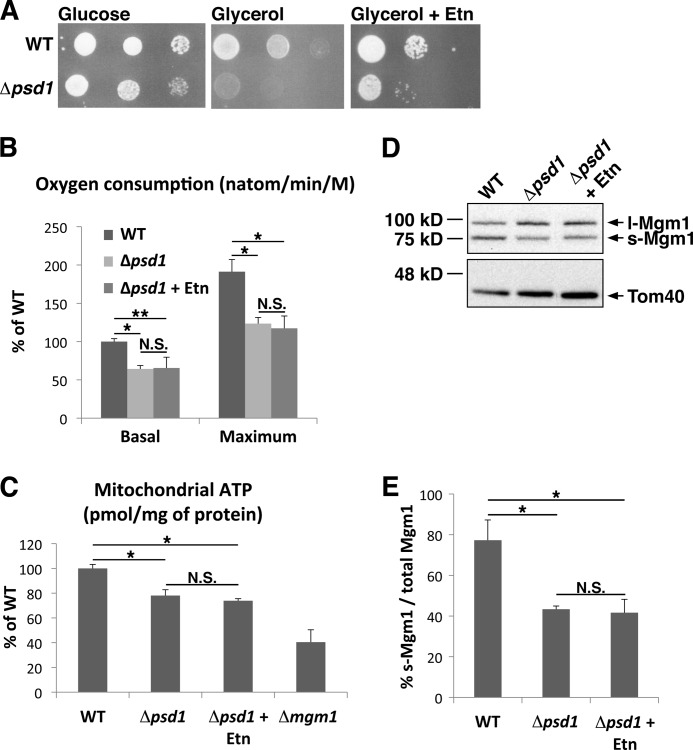
To better define the importance of Psd1 in mitochondrial biology, we determined whether Psd1 is required for normal mitochondrial homeostatic functions such as cell growth and mitochondrial bioenergetics. We analyzed cell growth under respiratory conditions, overall rate of oxidative phosphorylation, and mitochondrial ATP levels. Serial dilutions indicate that the Δpsd1 strain grows slower than the WT strain on glycerol, a nonfermentable carbon source that necessitates respiration (Fig. 4A). To more quantitatively determine the mitochondrial metabolic defect in the Δpsd1 strain, we analyzed oxidative phosphorylation by examining the rate of oxygen consumed during ethanol metabolism. Our data revealed that the basal rate of oxidative phosphorylation in the Δpsd1 strain is 50% that of the WT strain (Fig. 4B). The mitochondrial uncoupler, carbonylcyanide m-chlorophenylhydrazone was added to determine the maximum activity of the electron transport chain (23). The maximum rate of oxidative phosphorylation in the Δpsd1 strain is also 50% that of the WT strain (Fig. 4B). To determine the effect of impaired oxidative phosphorylation on energy production, a key function of mitochondria, we analyzed the amount of ATP in purified mitochondria. Δpsd1 mitochondria had ∼75% of WT mitochondrial ATP levels, indicating that Psd1 is required for the maintenance of mitochondrial ATP (Fig. 4C). Together, these results highlight the importance of Psd1, and likely the phospholipid PE, in regulating crucial mitochondrial components such as the electron transport chain to maintain cellular respiration and energy production.
FIGURE 4.
The Δpsd1 strain has defects in mitochondrial activity. A, serial dilutions of WT and Δpsd1 strains on glucose, glycerol, and glycerol with Etn. B, rate of oxidative phosphorylation indicated by the rate of oxygen consumed (nanoatom/minute per million cells) measured as described under “Experimental Procedures.” C, amount of ATP in purified mitochondria (pmol/mg of protein) measured using a luciferase assay. Δmgm1 is a respiratory-incompetent strain used as a negative control. D, Western blot analysis of l-Mgm1 and s-Mgm1 protein levels in WT, Δpsd1, and Δpsd1 cells grown in the presence of Etn. E, quantification of Western blots described in D. All values represent the means ± S.D. (error bars) of three independent experiments. *, p < 0.007; **, p < 0.02; N.S., not significant (p > 0.05).
Ethanolamine Cannot Rescue Δpsd1 Mitochondrial-specific Defects
In Saccharomyces cerevisiae, the major route of PE synthesis is by decarboxylation of phosphatidylserine (PS) by the mitochondrial-localized Psd1 and the secretory pathway-localized Psd2. Exogenously added ethanolamine (Etn) or Etn formed endogenously by lipid metabolism can also be used to synthesize PE via the Kennedy pathway (24). Because Psd1-dependent PS decarboxylation is the major route of PE synthesis, the loss of Psd1 results in significantly reduced mitochondrial and cellular PE. It was also reported that supplementing the Δpsd1 strain with exogenous Etn does not significantly increase mitochondrial PE content, indicating that exogenous PE is not efficiently transported to mitochondria (21). To directly demonstrate that the mitochondrial defects are a result of a reduction in mitochondrial PE, we cultured Δpsd1 cells in medium supplemented with Etn, which should increase extramitochondrial PE via the Kennedy pathway. The Δpsd1 glycerol growth defect is partially rescued in the presence of Etn (Fig. 4A). Interestingly, this rescue is independent of mitochondrial bioenergetics. The addition of Etn could not rescue the Δpsd1 oxidative phosphorylation or mitochondrial ATP defects (Fig. 4, B and C). These results strongly indicate that total cellular PE is crucial for growth and that mitochondrial-localized PE is required for efficient mitochondrial bioenergetics.
To assess whether mitochondrial PE is important for the maintenance of proper mitochondrial morphology, we analyzed the mitochondrial morphology of Δpsd1 cells expressing mtGFP grown in the presence of Etn as previously described. Under these conditions, 50% of Δpsd1 cells had an intermediate mitochondrial morphology. Mitochondria in these cells, although mostly tubular, were not connected in a network like those of WT cells (Fig. 1, A and B). This resembles the mitochondrial morphology of Δpsd1 cells cultured in rich medium, indicating that Psd1 is required for proper mitochondrial morphology, especially under more stringent, nutrient-limiting growth conditions.
Interestingly, it was previously observed that the loss of Psd1 results in altered ratios of l- and s-Mgm1 (25). This could suggest that the mitochondrial morphology defect in Δpsd1 cells is due to altered Mgm1 ratios. Thus, we determined whether the addition of Etn could alter Mgm1 protein ratios. Cells analyzed for their mitochondrial morphology were also harvested for Western blotting using Mgm1-specific antibodies. The ratio of l-Mgm1 and s-Mgm1 was not altered in the presence of Etn (Fig. 4, D and E), indicating that the rescue of mitochondrial fragmentation and aggregation in Δpsd1 is not a result of restoring Mgm1 ratios. The inability of Etn to fully restore the tubular mitochondrial network and Mgm1 ratios suggests that the imbalance of l- and s-Mgm1 protein levels contributes to the mitochondrial morphology defect in the Δpsd1 strain.
s*Mgm1 Can Rescue Mitochondrial Aggregation but Not the Glycerol Growth Defect in Δpsd1 Cells
A delicate balance of l- and s-Mgm1 is required for proper mitochondrial fusion. l-Mgm1 exerts a dominant negative effect; increased accumulation of l-Mgm1 results in impaired mitochondrial fusion (15). To address the possibility that the l- and s-Mgm1 protein imbalance contributes to the mitochondrial morphology defect in the Δpsd1 strain, we artificially increased the levels of s-Mgm1 by expressing a modified form of Mgm1 (s*Mgm1) on an exogenous plasmid (15). Expression of s*Mgm1 in the Δpsd1 strain markedly reduced mitochondrial aggregation and increased the number of cells with a tubular mitochondrial network (Fig. 5A). Culturing Δpsd1 cells expressing s*Mgm1 in Etn showed no further rescue of mitochondrial morphology (Fig. 5A). Although s*Mgm1 could rescue the mitochondrial network and aggregation, serial dilutions indicate that Δpsd1 cells expressing s*Mgm1 still exhibit impaired growth on glycerol (Fig. 5, B and C). These results indicate that s*Mgm1 could not restore mitochondrial activity in Δpsd1 despite restoring mitochondrial morphology.
FIGURE 5.
s*Mgm1 suppresses Δpsd1 mitochondrial aggregation. A, WT and Δpsd1 strains expressing mtGFP transformed with an empty vector (vector) or s*Mgm1 and cultured in the presence or absence of Etn. Mitochondrial morphology was quantified as in Fig. 1B. Quantification represents the mean ± S.D. (error bars) of three independent experiments. B, serial dilutions of WT and Δpsd1 cells with and without s*Mgm1 expression plasmid grown on glucose and glycerol. C, Western blot analysis indicating that s*Mgm1 is expressed.
Psd1 Regulates Alternative Topogenesis of Mgm1
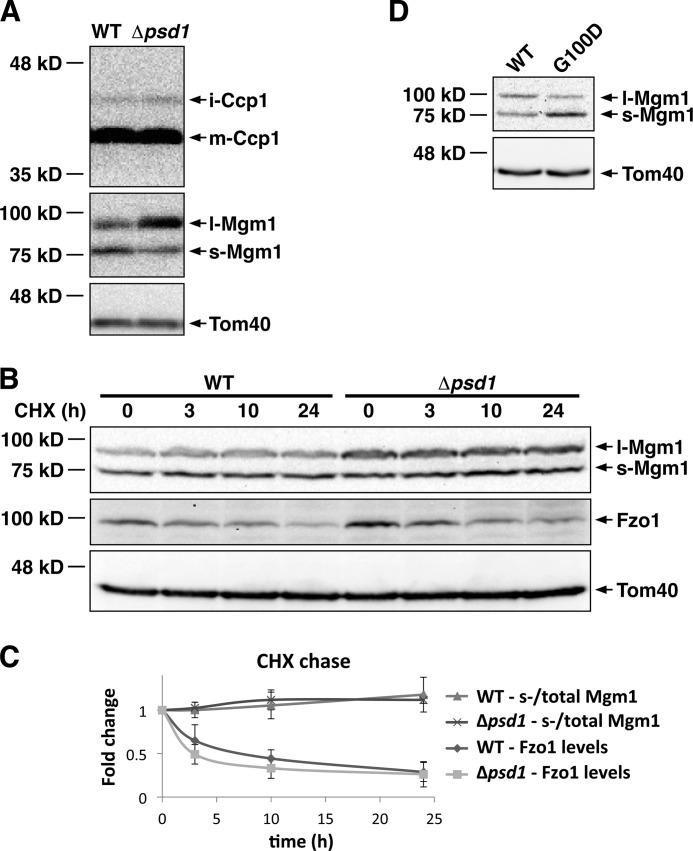
Our data indicate that the mitochondrial morphology defect in Δpsd1 cells is partly due to altered l- and s-Mgm1 protein levels. To define how Psd1 regulates Mgm1, we determined whether Psd1 regulates the biogenesis, proteolysis, and/or degradation of Mgm1. Mgm1 has two N-terminal hydrophobic regions. Insertion of the first hydrophobic region of FL-Mgm1 into the mitochondrial inner membrane results in the formation of l-Mgm1. In an ATP-dependent process, FL-Mgm1 can be inserted into the membrane via the second hydrophobic region, bypassing the first hydrophobic region (17). Subsequent proteolytic cleavage in the second hydrophobic region of Mgm1 by the rhomboid protease Rbd1/Pcp1 results in the formation of s-Mgm1. This alternative formation of l-Mgm1 and s-Mgm1 from FL-Mgm1 is known as Mgm1 alternative topogenesis (16, 26). It was previously shown that reducing the hydrophobicity of the first hydrophobic region allows Mgm1 to bypass alternative topogenesis, resulting in most of the Mgm1 protein being converted to s-Mgm1 (17). To determine whether Psd1 regulates the balance of l- and s-Mgm1 by regulating Rbd1, we analyzed the processing of Ccp1, another substrate of Rbd1. The processing of Ccp1 in the Δpsd1 strain was indistinguishable from that of the WT strain (Fig. 6A), indicating that Psd1 is not simply required for the activity of Rbd1 but specifically regulates Mgm1. To determine whether Psd1 regulates Mgm1 degradation, we performed a cycloheximide (CHX) chase. Consistently, we showed that the loss of Psd1 results in an increased l- to s-Mgm1 protein ratio (Fig. 6B). To determine whether this altered ratio is due to altered Mgm1 degradation, we analyzed the change in s-Mgm1 compared with total Mgm1 levels as a ratio during our CHX chase. A difference in the ratio between the WT and Δpsd1 strains would indicate that Psd1 regulates the degradation of l-Mgm1 and/or s-Mgm1. Protein ratios at the different time points were normalized to the starting protein ratio for each strain. Our analysis indicated no change in the ratio of s-Mgm1 compared with total Mgm1 levels over the course of 24 h during our CHX chase (Fig. 6, B and C). This result indicates that l- and s-Mgm1 are turned over at similar rates in the WT and Δpsd1 strains and that differential turnover of l- and s-Mgm1 is not the cause of altered l- and s-Mgm1 protein ratios in the Δpsd1 strain. As a positive control for the CHX chase, we also analyzed the degradation of Fzo1. In the WT strain, Fzo1 protein levels decreased over the course of the CHX chase, indicating that Fzo1 is rapidly degraded (Fig. 6, B and C), as previously observed (27). Interestingly, Fzo1 degradation is not impaired in the Δpsd1 strain. This is further evidence that Psd1 specifically regulates Mgm1 protein levels rather than globally regulating the stability of mitochondrial proteins. Because the formation of s-Mgm1 is ATP-dependent, our result that Psd1 is required for the maintenance of mitochondrial ATP levels (Fig. 4C) suggests that Psd1 regulates the ATP-dependent mechanism of s-Mgm1 biogenesis. To more definitively show that Psd1 regulates Mgm1 alternative topogenesis, we analyzed Δpsd1Δmgm1 cells expressing WT Mgm1 or Mgm1-G100D, a previously described point mutant of Mgm1 (17). The G100D point mutation results in reduced hydrophobicity in the first hydrophobic region of Mgm1, allowing it to bypass alternative topogenesis. This results in most of the Mgm1-G100D protein being converted to s-Mgm1 (17). Consistent with previous data, the Δpsd1Δmgm1 strain expressing a plasmid-borne copy of WT Mgm1 had less s-Mgm1 compared with l-Mgm1 (Fig. 6D). In contrast, the expression of Mgm1-G100D in Δpsd1Δmgm1 cells resulted in most of the Mgm1 protein being converted to s-Mgm1 (Fig. 6D). Taken together, our findings indicate that Psd1 regulates the balance of l- and s-Mgm1 by regulating Mgm1 alternative topogenesis, likely through its ATP-dependent mechanism.
FIGURE 6.
Psd1 regulates Mgm1 alternative topogenesis. A, Western blot analysis of WT and Δpsd1 strains expressing HA-tagged Ccp1. B, CHX chase performed for 24 h. WT and Δpsd1 cells were harvested at the indicated time points and analyzed by Western blotting using Mgm1, Fzo1, and Tom40 antisera. C, fold changes in s-Mgm1/total Mgm1 and Fzo1 protein levels as quantified by densitometry of the Western blots described in B. D, Western blot analysis of Δpsd1Δmgm1 cells expressing WT or the G100D point mutant of Mgm1. Values represent the mean ± S.D. of three independent experiments.
DISCUSSION
Mitochondrial dynamics have emerged as a critical aspect of mitochondrial biology, and they are implicated in several cellular outputs including apoptosis and autophagy. In the last decade, the protein machineries of membrane fusion and fission have been the focus of intense study, and the integral protein units that orchestrate these membrane dynamics have been well characterized. The next phase of discovery will be to understand the intricate mechanisms that regulate mitochondrial dynamics and how they are integrated into cellular signaling networks. Here, we have characterized the role of mitochondrial lipid metabolism in regulating overall mitochondrial biology, and more specifically membrane fusion.
Previous reports have indicated that Psd1 impacts mitochondrial morphology. We have identified the specific mitochondrial defects that arise when mitochondrial phospholipid composition is altered. A recent study that dramatically reduced both CL and PE demonstrated defects in mitochondrial membrane fusion and suggested overlapping roles of these phospholipids in mitochondrial activity (5). We have uncovered several mechanistic details of mitochondrial dysfunctions when only Psd1 is removed from mitochondria. Interestingly, most of the defects are hidden when cells are grown in rich medium, likely the result of compensatory lipid metabolic pathways that use the additional nutrients found in bacterial and yeast extracts present in rich media. Indeed, we have shown that mitochondria are tubular in Δpsd1 cells grown in rich medium, but are fragmented and aggregated in minimal medium (Fig. 1B). As shown by rescue experiments, mitochondrial aggregation in the Δpsd1 strain grown in minimal medium can be rescued by the addition of Etn (Fig. 1B). This strongly indicates that nutrients like Etn in the rich growth medium affect the mitochondrial morphology of Δpsd1 cells.
The ability of Etn to rescue mitochondrial aggregation but not the tubular mitochondrial network in the Δpsd1 strain could be due to increased OMM PE due to lipid transfer at mitochondrial-ER contact sites. It has recently been shown that an ERMES (ER-Mitochondria encounter structure) complex consisting of Mmm1, Mdm10, Mdm12, and Mdm34 tethers ER and mitochondria, facilitating phospholipid exchange between these organelles (28). It has also been shown that the secretory pathway-localized Psd2 and the Kennedy pathway contribute more PE to the OMM than to the IMM (19). This is further supported by our data indicating that IMM functions are not restored with Etn supplementation. Oxidative phosphorylation and mitochondrial ATP levels, which require a functional electron transport chain in the IMM, are not restored in the Δpsd1 strain in the presence of Etn (Fig. 4, B and C). Indeed, it has previously been shown that PE influences the activity of enzymes in the electron transport chain (29). This further stresses the importance of Psd1 in supplying mitochondrial PE to maintain proper mitochondrial bioenergetic activity. Curiously, although Etn supplementation could not rescue mitochondrial bioenergetics, it could moderately suppress the glycerol growth defect in the Δpsd1 strain (Fig. 4A). This result strongly indicates that lipid homeostasis not only affects mitochondrial functions, but other pathways required for proper cellular growth, bringing further attention to the importance of Psd1 in maintaining cellular and organellar lipid homeostasis.
In addition to a role in maintaining mitochondrial function, our study reveals that Psd1 maintains proper mitochondrial morphology by regulating mitochondrial fusion. Our data strongly indicate that the mitochondrial fusion defect in the Δpsd1 strain is due to both a reduced rate of lipid mixing from the altered biophysical properties of the mitochondrial membrane and also impaired Mgm1-driven mitochondrial fusion. Liposomes with phospholipid compositions similar to that of Δpsd1 mitochondria fuse to the same extent as those with compositions similar to that of WT mitochondria. However, their rate of fusion is only ∼25% that of WT (Fig. 3, A and B). Alterations in the lipid composition can affect many properties of the lipid membrane that are believed to be important for membrane fusion. PE is known to generate negative membrane curvature, increase lipid-packing stress, and induce the formation of hexagonal phases-all of which are properties that enhance membrane fusion (30–32). Here, we show that these alterations to the biophysical properties of liposomes with a lipid composition similar to that of the mitochondrial membrane reduce their fusion kinetics. However, this in vitro experiment was performed in the absence of proteins and a second bilayer, factors that promote mitochondrial fusion. Nevertheless, our result suggests that the lipid composition is unlikely to regulate the extent of fusion, but might regulate the kinetics of fusion. To our knowledge, this is the first indication that the kinetics of mitochondrial fusion could be regulated by its phospholipid composition.
The ability of s*Mgm1 to significantly reduce mitochondrial aggregation in the Δpsd1 strain strongly indicates that Psd1 regulates mitochondrial morphology in part by regulating alternative topogenesis and the biogenesis of Mgm1. Although we might have expected the combination of s*Mgm1 and Etn supplementation to fully rescue the mitochondrial morphology defect in the Δpsd1 strain, we observed no significant improvement in the presence of Etn when Δpsd1 cells expressed s*Mgm1 (Fig. 5A). Although s*Mgm1 expression rescued the imbalance of l-Mgm1 and s-Mgm1 in Δpsd1 cells, it could not restore the phospholipid balance of the mitochondrial membrane. This is compelling evidence that Psd1 plays many nonoverlapping roles in mitochondrial regulation, all of which are critical in mitochondrial biology and further substantiates the concept that the phospholipid composition of the IMM is critical for proper mitochondrial activity.
Interestingly, we observed differing severities of the Δpsd1 mitochondrial morphology defect. When the cells expressed the HIS3 gene (Fig. 1B), the morphology defect was more severe than when they expressed both the HIS3 and LEU2 genes (Fig. 5A). Only 50% of Δpsd1 cells had fragmented and aggregated mitochondria when expressing the HIS3 and LEU2 genes compared with 85% when expressing only the HIS3 gene (Figs. 5A and 1B, respectively). This is a strong indication that lipid homeostasis is closely linked to amino acid biosynthesis. Indeed, leucine was previously identified as a precursor of phospholipids such as PS, PE, and phosphatidylcholine (33). It is possible that the ability to synthesize leucine allowed for the conversion of some leucine to PE, compensating for the reduction in PE levels in the Δpsd1 strain.
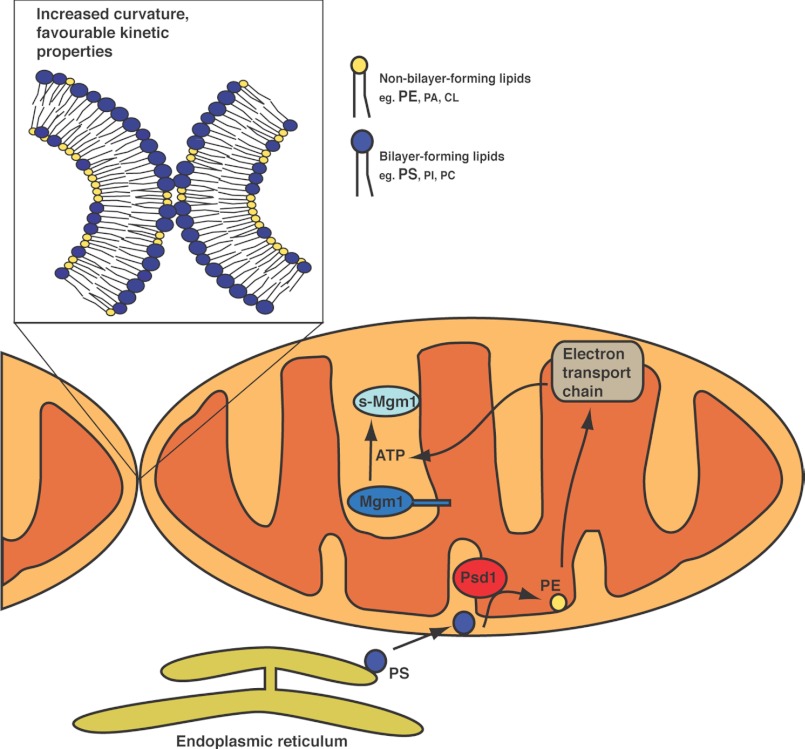
Our current study reveals that Psd1 regulates mitochondrial morphology by enhancing the rate of lipid mixing during mitochondrial fusion. We also show that Psd1 plays crucial roles in maintaining mitochondrial functions such as oxidative phosphorylation and maintaining ATP levels. In addition, impaired Mgm1 alternative topogenesis in Δpsd1 cells impedes s-Mgm1 biogenesis, further exacerbating the mitochondrial fusion defect (Fig. 7). Our work highlights the dual function of Psd1 in regulating mitochondrial fusion by enhancing lipid kinetics and the biogenesis of a mitochondrial fusion protein.
FIGURE 7.
Model of Psd1-dependent mitochondrial regulation. PE generates negative membrane curvature, promoting fusion. In addition, the headgroup of PE provides favorable kinetic conditions. Because PE is known to regulate complexes of the electron transport chain, energy production is impaired with reduced PE, further contributing to the mitochondrial fusion defect by impairing s-Mgm1 protein biosynthesis.
Although studies in the last decade have focused on the protein machinery of mitochondrial dynamics, several recent reports have brought lipid homeostasis to the forefront of mitochondrial biology. Our work and work of others have identified critical roles of phospholipids in maintaining cellular and organellar competence, highlighting their critical biological importance.
Acknowledgments
We thank Dr. Benedikt Westermann, Dr. Andreas Reichert, Dr. Jodi Nunnari, and Dr. Thomas Langer for reagents and all members of the McQuibban laboratory for helpful discussions.
This work was supported by an operating grant from the Canadian Institutes of Health Research.
- CL
- cardiolipin
- CHX
- cycloheximide
- Etn
- ethanolamine
- FL-
- full-length
- IMM
- inner mitochondrial membrane
- l-
- long
- OMM
- outer mitochondrial membrane
- PE
- phosphatidylethanolamine
- PS
- phosphatidylserine
- s-
- short.
REFERENCES
- 1. Choi S. Y., Huang P., Jenkins G. M., Chan D. C., Schiller J., Frohman M. A. (2006) A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat. Cell Biol. 8, 1255–1262 [DOI] [PubMed] [Google Scholar]
- 2. Tamura Y., Endo T., Iijima M., Sesaki H. (2009) Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J. Cell Biol. 185, 1029–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sakamoto T., Inoue T., Otomo Y., Yokomori N., Ohno M., Arai H., Nakagawa Y. (2012) Deficiency of cardiolipin synthase causes abnormal mitochondrial function and morphology in germ cells of Caenorhabditis elegans. J. Biol. Chem. 287, 4590–4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kuroda T., Tani M., Moriguchi A., Tokunaga S., Higuchi T., Kitada S., Kuge O. (2011) FMP30 is required for the maintenance of a normal cardiolipin level and mitochondrial morphology in the absence of mitochondrial phosphatidylethanolamine synthesis. Mol. Microbiol. 80, 248–265 [DOI] [PubMed] [Google Scholar]
- 5. Joshi A. S., Thompson M. N., Fei N., Hüttemann M., Greenberg M. L. (2012) Cardiolipin and mitochondrial phosphatidylethanolamine have overlapping functions in mitochondrial fusion in Saccharomyces cerevisiae. J. Biol. Chem. 287, 17589–17597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Steenbergen R., Nanowski T. S., Beigneux A., Kulinski A., Young S. G., Vance J. E. (2005) Disruption of the phosphatidylserine decarboxylase gene in mice causes embryonic lethality and mitochondrial defects. J. Biol. Chem. 280, 40032–40040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van den Brink-van der Laan E., Killian J. A., de Kruijff B. (2004) Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim. Biophys. Acta 1666, 275–288 [DOI] [PubMed] [Google Scholar]
- 8. Churchward M. A., Rogasevskaia T., Brandman D. M., Khosravani H., Nava P., Atkinson J. K., Coorssen J. R. (2008) Specific lipids supply critical negative spontaneous curvature–an essential component of native Ca2+-triggered membrane fusion. Biophys. J. 94, 3976–3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hermann G. J., Thatcher J. W., Mills J. P., Hales K. G., Fuller M. T., Nunnari J., Shaw J. M. (1998) Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143, 359–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eura Y., Ishihara N., Yokota S., Mihara K. (2003) Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J. Biochem. 134, 333–344 [DOI] [PubMed] [Google Scholar]
- 11. Sesaki H., Southard S. M., Yaffe M. P., Jensen R. E. (2003) Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane. Mol. Biol. Cell 14, 2342–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cipolat S., Martins de Brito O., Dal Zilio B., Scorrano L. (2004) OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. U.S.A. 101, 15927–15932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sesaki H., Jensen R. E. (2004) Ugo1p links the Fzo1p and Mgm1p GTPases for mitochondrial fusion. J. Biol. Chem. 279, 28298–28303 [DOI] [PubMed] [Google Scholar]
- 14. Herlan M., Vogel F., Bornhovd C., Neupert W., Reichert A. S. (2003) Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J. Biol. Chem. 278, 27781–27788 [DOI] [PubMed] [Google Scholar]
- 15. Zick M., Duvezin-Caubet S., Schäfer A., Vogel F., Neupert W., Reichert A. S. (2009) Distinct roles of the two isoforms of the dynamin-like GTPase Mgm1 in mitochondrial fusion. FEBS Lett. 583, 2237–2243 [DOI] [PubMed] [Google Scholar]
- 16. McQuibban G. A., Saurya S., Freeman M. (2003) Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature 423, 537–541 [DOI] [PubMed] [Google Scholar]
- 17. Herlan M., Bornhövd C., Hell K., Neupert W., Reichert A. S. (2004) Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor. J. Cell Biol. 165, 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Westermann B., Neupert W. (2000) Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16, 1421–1427 [DOI] [PubMed] [Google Scholar]
- 19. Bürgermeister M., Birner-Grünberger R., Nebauer R., Daum G. (2004) Contribution of different pathways to the supply of phosphatidylethanolamine and phosphatidylcholine to mitochondrial membranes of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1686, 161–168 [DOI] [PubMed] [Google Scholar]
- 20. Gregg C., Kyryakov P., Titorenko V. I. (2009) Purification of mitochondria from yeast cells. J. Vis. Exp. e1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Birner R., Bürgermeister M., Schneiter R., Daum G. (2001) Roles of phosphatidylethanolamine and of its several biosynthetic pathways in Saccharomyces cerevisiae. Mol. Biol. Cell 12, 997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dimmer K. S., Fritz S., Fuchs F., Messerschmitt M., Weinbach N., Neupert W., Westermann B. (2002) Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen H. K., Ji Z. S., Dodson S. E., Miranda R. D., Rosenblum C. I., Reynolds I. J., Freedman S. B., Weisgraber K. H., Huang Y., Mahley R. W. (2011) Apolipoprotein E4 domain interaction mediates detrimental effects on mitochondria and is a potential therapeutic target for Alzheimer disease. J. Biol. Chem. 286, 5215–5221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Daum G., Lees N. D., Bard M., Dickson R. (1998) Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast 14, 1471–1510 [DOI] [PubMed] [Google Scholar]
- 25. Osman C., Haag M., Potting C., Rodenfels J., Dip P. V., Wieland F. T., Brügger B., Westermann B., Langer T. (2009) The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J. Cell Biol. 184, 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sesaki H., Southard S. M., Hobbs A. E., Jensen R. E. (2003) Cells lacking Pcp1p/Ugo2p, a rhomboid-like protease required for Mgm1p processing, lose mtDNA and mitochondrial structure in a Dnm1p-dependent manner, but remain competent for mitochondrial fusion. Biochem. Biophys. Res. Commun. 308, 276–283 [DOI] [PubMed] [Google Scholar]
- 27. Fritz S., Weinbach N., Westermann B. (2003) Mdm30 is an F-box protein required for maintenance of fusion-competent mitochondria in yeast. Mol. Biol. Cell 14, 2303–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kornmann B., Currie E., Collins S. R., Schuldiner M., Nunnari J., Weissman J. S., Walter P. (2009) An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Daum G. (1985) Lipids of mitochondria. Biochim. Biophys. Acta 822, 1–42 [DOI] [PubMed] [Google Scholar]
- 30. Epand R. M., Fuller N., Rand R. P. (1996) Role of the position of unsaturation on the phase behavior and intrinsic curvature of phosphatidylethanolamines. Biophys. J. 71, 1806–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang Q., Guo Y., Li L., Hui S. W. (1997) Effects of lipid headgroup and packing stress on poly(ethylene glycol)-induced phospholipid vesicle aggregation and fusion. Biophys. J. 73, 277–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Siegel D. P., Epand R. M. (1997) The mechanism of lamellar-to-inverted hexagonal phase transitions in phosphatidylethanolamine: implications for membrane fusion mechanisms. Biophys. J. 73, 3089–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ramsey R. B. (1976) Leucine and d-3-hydroxybutyrate as lipid precursors in developing rat spinal cord and liver. Biochem. J. 158, 501–504 [DOI] [PMC free article] [PubMed] [Google Scholar]