Background: Negative feedback regulation of insulin signaling involves ubiquitin-dependent degradation of insulin receptor substrate 1 (IRS1).
Results: Cullin-RING E3 ubiquitin ligase 7 (CRL7) mediates the ubiquitination of IRS1 in hyperphosphorylated form.
Conclusion: Multisite IRS1 phosphorylation triggers interactions with CRL7 for ubiquitin modification.
Significance: Insulin signaling is self-restrained when its downstream effector kinases are hyperactivated to trigger the negative feedback inhibition.
Keywords: Insulin, Proteasome, Signaling, Ubiquitin Ligase, Ubiquitination
Abstract
Hyperactivation of mechanistic target of rapamycin complex 1 (mTORC1) and its effector kinase S6 kinase 1 (S6K1) is known to trigger multisite seryl phosphorylation of insulin receptor substrate 1 (IRS1), leading to its ubiquitination and degradation. This negative feedback inhibition functions to restrain PI3K activity and plays critical roles in the pathogenesis of cancer and type II diabetes. Recent work has implicated a role for cullin-RING E3 ubiquitin ligase 7 (CRL7) in targeting IRS1 for mTORC1/S6K1-dependent degradation. In the present study we have employed both cell-based degradation and reconstituted ubiquitination approaches to define molecular features associated with IRS1 critical for CRL7-mediated ubiquitination and degradation. We have mapped IRS1 degradation signal sequence to its N-terminal 574 amino acid residues, of which the integrity of Ser-307/Ser-312 and Ser-527, each constituting a S6K1 phosphorylation consensus site, was indispensible for supporting CRL7-forced degradation. In vitro, S6K1 was able to support the ubiquitination of bacterially expressed IRS1 N-terminal fragment by CRL7 but at low levels. In contrast, CRL7 supported efficient ubiquitination of IRS1 N-terminal fragment in hyperphosphorylated form, which was isolated from infected insect cells, suggesting requirement of additional phosphorylation by kinases yet to be identified. Finally, removal of IRS1 amino acids 1–260 led to substantial reduction of ubiquitination efficiency, suggesting a role for this region in mediating productive interactions with CRL7. The requirement of multisite phosphorylation and the N terminus of IRS1 for its turnover may ensure that complete IRS1 degradation occurs only when mTORC1 and S6K1 reach exceedingly high levels.
Introduction
In mammals, growth regulation is mediated through a signal integrator known as the protein kinase mechanistic target of rapamycin (mTOR)3 (1, 2). mTOR comprises two distinct complexes named mTORC1 and mTORC2. mTORC1 integrates signals from at least five major inputs including nutrients, growth factors, energy, oxygen, and stress. Depending on the nutritional state of the organism, growth factors, such as insulin, signal to mTORC1 by initiating binding to their receptors. Insulin, or insulin-like growth factor 1, receptors are tyrosine kinases, which, upon ligand binding, phosphorylate the insulin receptor substrate (IRS), such as IRS1 at multiple tyrosine residues. The resulting IRS1 phosphotyrosines provide docking sites capable of recruiting a set of key Src homology 2-containing signaling proteins that include PI3K and Grb2 (growth factor receptor-bound protein 2), thus activating the downstream Akt (via PI3K) and RAS (through Grb2) pathways. In the Akt cascade, it is thought that the activated Akt inhibits the TSC1/2 (tuberous sclerosis 1/2) complex, thereby liberating the small G-protein Rheb (Ras homologue enriched in brain), which in turn activates mTORC1 and its downstream effector kinase S6 kinase 1 (S6K1), leading to elevated ribosome biogenesis and cell growth (reviewed in Refs. 3–5).
Optimal mTORC1 signaling requires the mTORC1/S6K1-IRS1 negative feedback inhibition that attenuates the strength or duration of PI3K activity (4). This inhibitory loop is triggered by hyperactivated mTORC1/S6K1 that catalyze multisite IRS1 seryl phosphorylation, which suppresses the ability of IRS1 to interact with the insulin/insulin-like growth factor 1 receptors and promotes its ubiquitin-dependent proteasomal degradation (6). The mTORC1/S6K-IRS1 feedback loop appears to directly impact the pathogenesis of diabetes and cancer. It was shown that overfeeding causes hyperactivation of mTORC1 that triggers the mTORC1/S6K1-IRS1 feedback loop, thereby leading to insulin desensitization (7). Down-regulation of IRS1 activity suppresses Akt, causing reduction of glucose uptake and glycogen synthesis in liver and muscle, as well as the increase of gluconeogenesis and glucose release by the liver. Thus, the mTORC1/S6K1-IRS1 feedback worsens hyperglycaemia and hyperinsulinaemia, which are clinical conditions resulting from excess nutrients. In further support of the role of the mTORC1/S6K1-IRS1 feedback loop in the pathogenesis of type 2 diabetes, S6K1-deficient mice displayed enhanced insulin sensitivity when chronically maintained on a high fat diet (7).
Previous mouse studies have elucidated a role for the mTORC1/S6K1-IRS1 negative feedback loop in cancer suppression (8). It was shown that TSC2+/− mice develop relatively benign hemangiomas because of sporadic loss of the functional Tsc2 allele. The lack of malignant tumor development is resulted from inactive Akt, because TSC2 down-regulation leads to hyperactivation of mTORC1/S6K1, which inactivates IRS1 and hence PI3K. Indeed, circumventing the mTORC1/S6K1-IRS1 feedback loop by reduction of PTEN (phosphatase and tensin homologue) activity results in enhanced Akt activation, as well as more frequent and aggressive hemangiomas (8). Therefore, it was proposed that the mTORC1/S6K1-IRS1 negative feedback loop plays a critical role in restraining PI3K activity, thereby halting the progression to malignancy. In addition, the mTORC1/S6K1-IRS1 feedback loop appears to limit the efficacy of rapamycin, a potent inhibitor of mTORC1, as an anti-cancer drug in clinical trials. Rapamycin inhibits mTORC1 and hence the mTROC1/Sk61-IRS1 feedback loop, resulting in severe up-regulation of PI3K. Thus, by shutting down the mTORC1/S6K1-IRS1 feedback loop, the rapamycin-based therapeutic approaches may elevate PI3K activity, which provides important pro-survival and proliferative signals through Akt (9), partly explaining why rapamycin is cytostatic but not cytotoxic in many tumors.
Haruta et al. (10) initially observed proteolytic turnover of IRS1 during prolonged exposure to insulin. The IRS1 down-regulation requires PI3K and mTORC1, because it is sensitive to treatment by wortmannin, a PI3K inhibitor, and by rapamycin, the mTORC1 inhibitor. Subsequent studies have since confirmed these results and have further determined that the turnover of IRS1 requires the 26 S proteasome and ubiquitination activity, respectively (6). Furthermore, the integrity of Ser-312, a target of mTORC1, was shown to mediate, at least in part, the degradation of human IRS1 (11). Our recent work has suggested a role for Cullin-RING E3 ubiquitin ligase 7 (CRL7) in targeting IRS1 for ubiquitin-dependent degradation (12).
CRL7 is a member of the CRLs (Cullin-RING finger E3 ligases), which comprise the largest E3 family responsible for directing the polyubiquitination of substrate proteins, thereby leading to their eventual degradation by the 26 S proteasome (13). CRL7 contains cullin 7 (CUL7) as a molecular scaffold, the F-box protein Fbw8 as a substrate receptor, Skp1, and the ROC1 (Rbx1) RING finger protein. In the CRL7 complex, CUL7 assembles both an Skp1·Fbw8 heterodimeric substrate-targeting module and a ROC1 RING-based ubiquitin core ligase. It is thought that the organization of the CRL7 subunits places Fbw8 within the proximity of ROC1, which recruits an E2-conjugating enzyme. Consequently, a substrate, once bound to Fbw8, is positioned optimally for accepting an ubiquitin moiety in an E2-catalyzed transfer reaction. Cormier-Daire and co-workers (14, 15) have found a large panel of cul7 germ line mutations in patients with the 3-M syndrome, which is characterized by pre- and post-natal growth retardation. In addition, both the cul7 (16) and fbw8 (17) null mice exhibit intrauterine growth retardation. Taken together, the genetic evidence has strongly suggested a pivotal role for CRL7 in growth control.
It has been shown that Fbw8 binds to IRS1 and promotes its ubiquitination and proteasomal degradation and that inactivation/deletion of Fbw8 and CUL7, respectively, accumulated IRS1 (12). Moreover, Fbw8-induced degradation of IRS1 was dependent upon mTORC1 activity. In addition, embryonic fibroblasts of cul7−/− mice were found to accumulate IRS1 and exhibit increased activation of IRS1 downstream pathways Akt and MEK/ERK. These results have led to a proposal that CRL7 participates in the mTORC1/S6K1-IRS1 negative feedback control by targeting IRS1 for degradation. In the present study, we sought to understand the IRS1 molecular determinants that dictate its destruction by CRL7.
EXPERIMENTAL PROCEDURES
Plasmids
We constructed plasmids allowing expression in mammalian cells of human IRS1 regions starting with varying N-terminal sites and ending at amino acid position 574 using the previously generated pcDNA3.1-V5-IRS1–1-574 (12) as template. The primers used for PCR amplification of various IRS1 sequences are: 114–574, forward primer, CACCCACAACCGTGCTAAG, and reverse primer, GCGGGTGGGCACGAACTAGGAGTGCCTGTGTCC; 261–574, forward primer, CACCAGTGATGAGTTCCGC, and reverse primer, GCGGGTGGGCACGAACTAGGAGTGCCTGTGTCC; and 300–574, forward primer, CACCACCCGCCGATCACG, and reverse primer, GCGGGTGGGCACGAACTAGGAGTGCCTGTGTCC. Various truncations were first cloned into pENTR entry vector (Invitrogen), followed by TOPO cloning into the destination vector pcDNA3.1/nV5-DEST (Invitrogen) according to the manufacturer's instructions.
To generate human IRS1 1–574 point mutants, site-direct mutagenesis was performed with pcDNA3.1-V5-IRS1–1-574 as template, using the QuikChange® site-directed mutagenesis kit (Stratagene) and sets of primers as follows: V5-IRS1–1-574S307A/S312A, TCACGCACTGAGGCCATCACCGCCACCGCCCCGGCCAGCATG-5′, and CATGCTGGCCGGGGCGGTGGCGGTGATGGCCTCAGTGCGTGA-3′; and V5-IRS1–1-574S527A, CGAAAGAGAACTCACGCGGCAGGCACATCCCCT-5′ and AGGGGATGTGCCTGCCGCGTGAGTTCTCTTTCG-3′. V5-IRS-1S307A/S312AS527A was generated by performing mutagenesis reaction with V5-IRS-1S307A/S312A as template, using the set of primers for the construction of V5-IRS-1S527A. To create pGEX-4T3-GST-IRS1 1–574, IRS1 1–574 sequence was inserted into the multicloning sites on the pGEX-4T3 vector.
To construct bacmid DNA expressing GST-IRS1 1–574 or GST-IRS1–261-574, the respective DNA fragment encoding either of these proteins was amplified by PCR using pcDNA3.1-V5-IRS1–1-574 or pcDNA3.1-V5-IRS1 261–574 as template. The PCR primer sequences, designed to include the BamHI restriction site followed by the Kozak sequence (ACC) at 5′ end and the EcoRI restriction site at the 3′ end, were: ACCATCGGGCGCGGATCCACCATGTCCCCTATACTAGGT-5′ and CCGGAATTCCTAGGAGTGCCTGTGTCCC-3′. The resulting PCR product was then cloned into the BamHI and EcoRI predigested pFast-Bac1 vector (Bac-to-Bac baculovirus expression system; Invitrogen), creating pFast-GST-IRS1–1-574 or pFast-GST-IRS1–261-574. The recombinant pFast-GST-IRS1–1-574 or pFast-GST-IRS1–261-574 was transformed into competent DH10Bac cells where transposition takes place. The bacmid DNA containing GST-IRS1–1-574 or GST-IRS1–261-574 was isolated following the manufacturer's instructions. All of the constructs were verified by DNA sequencing.
Baculovirus
To prepare baculovirus expressing GST-IRS1–1-574 or GST-IRS1–261-574, Sf9 cells (9 × 105 cells in one well of a 6-well plate) were transfected with recombinant bacmid DNA pFast-GST-IRS1–1-574 or pFast-GST-IRS1–261-574 (2 μg each) using Cellfectin reagent (Invitrogen). A high titer viral stock was generated after three rounds of amplification as per the manufacturer's instructions (Invitrogen).
Cell Transfection and Extraction
Human embryonic kidney cells (HEK293) were grown on 2 ml/plate DMEM, 10% FBS, and 1% antibiotic-antimycotic (Invitrogen). DNA(s) was transfected up to 1 μg/plate using Lipofectamine 2000 (Invitrogen). To harvest cells at 48 h post-transfection, the plates are washed with 1 ml of PBS, and the cells were pelleted at 180 × g for 5 min using a Beckman CS-6KR centrifuge at 4 °C. The cell pellets were resuspended in Nonidet P-40 lysis buffer (20 mm Hepes-KOH, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA, 1 mm DTT, 1 mm PMSF, 12.5 mm NaF, 1 mm Na3VO4, 400 μm phenylarsine oxide, 10 μg/ml antipain, and 10 μg/ml leupeptin) in 0.2 ml/plate, and the resulting suspension was sonicated (seven repetitive 20-s treatments). The mixture was agitated for 30 min at 4 °C followed by centrifugation (at 100,000 × g at 4 °C for 30 min). The cleared extracts were used for immunoprecipitation/immunoblot analyses.
Generation of Stable Cell Lines Expressing FLAG-CUL7 and Myc-Fbw8 (293C7F8)
Generation of stable cell lines expressing FLAG-CUL7 and Myc-Fbw8 (293C7F8) was carried out using a previously published protocol (18). HEK293 cells, plated at a density of 2 × 106 cells/100-mm plate and maintained in 10 ml of the growth medium (DMEM supplemented with 10% FBS and 100 unit/ml of penicillin/streptomycin (Invitrogen), were co-transfected with a plasmid expressing the Geneticin selection marker, pcDNA3.1-FLAG-CUL7 and pCR3.1-Myc-Fbw8, using Lipofectamine 2000 (Invitrogen). At 48 h post-transfection, the cells were diluted at various ratios (1:5, 1:50, and 1:500) and plated onto 15-mm plates followed by the addition of the selection medium (25 ml/plate) containing the growth medium plus Geneticin (700 μg/ml; Invitrogen). The selection medium was changed daily for the first 3–5 days after the Geneticin treatment to remove dead cells and then every third day for the next 2 weeks. Well isolated colonies were picked 10–15 days using sterile cloning discs (Scienceware) presoaked in trypsin-EDTA (Invitrogen). Each disc was then placed into an individual well, containing the selection medium, within a 24-well chamber. 24 h later, the discs were removed, and the individual clones were allowed to proliferate in the selection medium. Protein expression was determined by immunoprecipitation/immunoblot analysis using extracts prepared from one 60-mm plate worth of cells.
Protein Isolation
Isolation of B-GST-IRS1 1–574
To prepare bacterially expressed GST-IRS1–1-574 (B-GST-IRS1–1-574), pGEX-GST-IRS1–1-574 was transformed into the pJY2 (Affiniti)-expressing-BL21 (DE3) cells and grown in Luria Broth (500 ml) with 0.4% glucose in the presence of ampicillin and chloramphenicol at 37 °C. When the optical density at 600 nm reached 0.4, the cultures were induced with 0.8 mm isopropyl-1-thio-β-d-galactopyranoside at 37 °C for 3 h. The cells were pelleted at 5,000 × g for 15 min at 4 °C followed by suspension in culture volume with lysis buffer (50 mm Tris-HCl, pH 8.0, 1% Triton X-100, 0.5 m NaCl, 10 mm EDTA, 10 mm EGTA, 10% glycerol, 0.1 mm PMSF, 0.4 μg/ml antipain, 0.2 μg/ml leupeptin, and 5 mm DTT). The suspension was then sonicated (four repetitive 20-s treatments) and centrifuged at 17,000 rpm in an SS-34 rotor for 30 min at 4 °C. Soluble extracts (20 ml) were mixed with glutathione-S-Sepharose beads (3 ml) for 2 h at 4 °C. The beads were then packed into a column and washed consecutively with buffer A (25 mm Tris-HCl, pH 7.5, 1 mm EDTA, 0.01% Nonidet P-40, 10% glycerol, 1 mm DTT, 0.2 μg/ml leupeptin, 0.2 μg/ml antipain, and 1 mm PMSF) plus 0.5 m NaCl (30 ml) and buffer A plus 0.1 m NaCl (15 ml). Bound protein was eluted with 15 ml of 20 mm reduced glutathione contained in buffer A plus 0.1 m NaCl. Following dialysis with buffer A plus 0.1 m NaCl, GST-IRS1–1-574 was then concentrated using centrifugal filters (Amicon-Millipore). The yield was 3.5 mg (7 mg/ml).
Isolation of BI-GST-IRS1 1–574 and BI-GST-IRS1 261–574
To prepare insect cell-expressed GST-IRS1 1–574 (BI-GST-IRS1 1–574) or GST-IRS1 261–574 (BI-GST-IRS1 261–574), Hive Five cells (3.25 × 107) were infected (multiplicity of infection of 5) with each baculovirus. At 48 h post-infection, the cells were harvested, washed once with cold PBS, and resuspended in 2 ml/flask of Nonidet P-40 lysis buffer (20 mm Hepes, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA, 1 mm DTT, 1 mm PMSF, 12.5 mm NaF, 1 mm Na3VO4, 400 μm phenylarsine oxide, 10 μg/ml antipain, and 10 μg/ml leupeptin). The resuspension was centrifuged at 15,000 rpm for 30min at 4 °C. Soluble extracts were mixed with glutathione-S-Sepharose beads (0.2 ml for 1.6 ml of supernatant from eight flasks). Purification was carried out as described above. The yields were 170 and 296 μg (1.7 and 2.96 mg/ml), respectively.
Isolation of CRL7
CRL7 was isolated from extracts derived from forty-four 150-mm plate worth of 293C7F8 cells (16CF) and maintained in buffer RSB (10 mm Tris-HCl, pH 7.4, 10 mm NaCl, 0.5% Nonidet P-40, 5 mm EDTA, 5 mm EGTA, 1 mm PMSF, 10 μg/ml antipain, and 10 μg/ml leupeptin). The mixture was then sonicated (three repetitive 20-s treatment), followed by the addition of buffer LSB (20 mm Tris-HCl, pH 7.4, 1 m NaCl, 0.5% Nonidet P-40, 5 mm EDTA, 5 mm EGTA, 1 mm PMSF, 10 μg/ml antipain, and 10 μg/ml leupeptin). The mixture was then rotated for 1 h at 4 °C, followed by ultracentrifugation at 37,000 rpm for 30min at 4 °C. 10 ml of resulting supernatant (∼5 mg of protein/ml) were adsorbed to M2-agarose beads (0.5 ml; Sigma) by rotating the mixture at 4 °C for 14–16 h. The resulting beads were packed into a column, which was washed with 100 ml of wash buffer (15 mm Tris-HCl, pH 7.4, 500 mm NaCl, 0.5% Nonidet P-40, 5 mm EDTA, 5 mm EGTA, 1 mm PMSF, 10 μg/ml antipain, and 10 μg/ml leupeptin. Bound proteins were eluted with FLAG peptide (1 mg/ml) in Buffer A plus 0.1 m NaCl, for three consecutive times, 1.5 ml/time. The pooled elutes were concentrated to ∼0.2 ml using centrifugal filters (Millipore).
In Vitro Analysis
For in vitro phosphorylation described in Fig. 3B, B-GST-IRS1–1-574 (7.7 pmol) was incubated with S6K1T389E/PDK (0.3 μg) at 37 °C for 45min in a reaction mixture (20 μl) containing 50 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 0.5 mm DTT, 2 mm ATP, 0.1 mg/ml BSA, 2 mm NaF, and 10 nm okadaic acid. For in vitro dephosphorylation described in Figs. 4A and 5A, BI-GST-IRS1–1-574 (10 pmol) or BI-GST-IRS1–261-574 (20 pmol) was incubated with λ phosphatase (New England BioLabs) at concentration of 0.1, 1, or 10 ng in a reaction mixture (20 μl) containing λ phosphatase buffer (New England BioLabs) at 37 °C for 30 min.
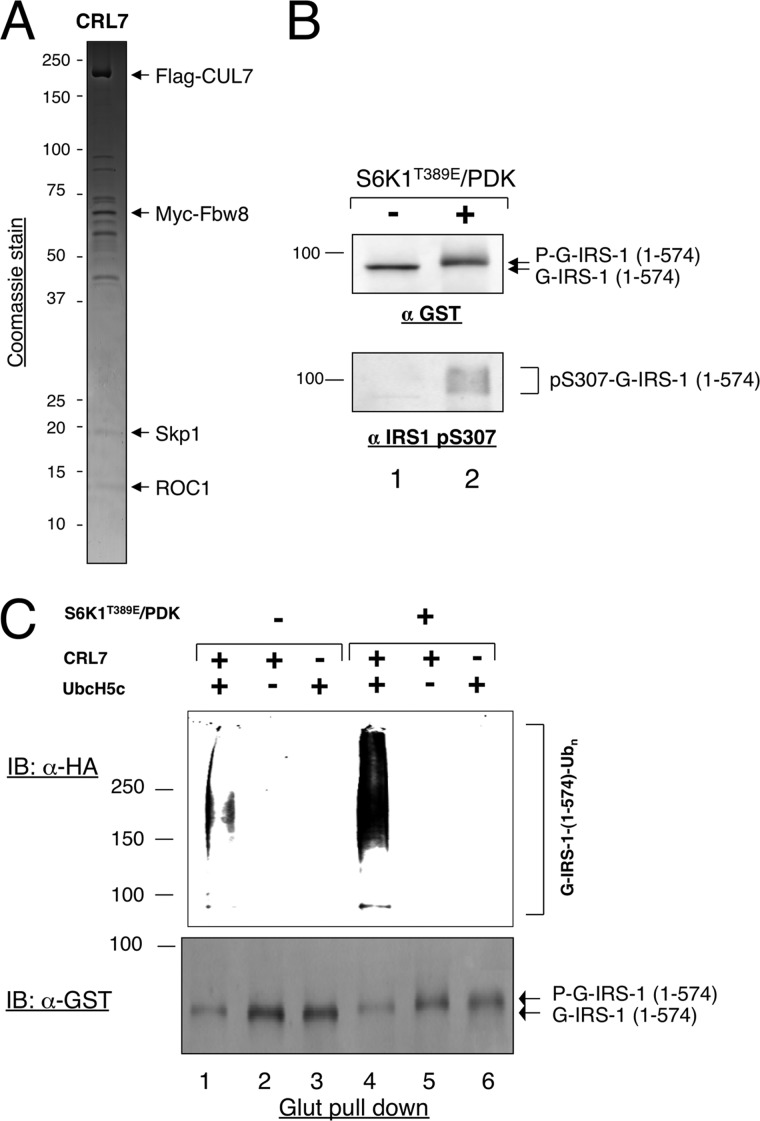
FIGURE 3.
S6K1 stimulates the ubiquitination of IRS1 1–574 by CRL7. A, Coomassie stain analysis of the affinity-purified CRL7. An aliquot (∼3 pmol) of purified CRL7 was subject to SDS-PAGE, followed by Coomassie staining. The CRL7 components are indicated. B, phosphorylation of IRS-1 1–574 by S6K1. Phosphorylation reaction was carried out as described under “Experimental Procedures.” The reaction products were analyzed by immunoblot (IB) analysis using indicated antibodies. C, reconstitution of in vitro polyubiquitination of IRS-1 1–574 by CRL7. Polyubiquitination of IRS-1 1–574 was reconstituted, in the presence or absence of S6K1 treatment, as described under “Experimental Procedures.” Immunoblot analysis with indicated antibodies was performed to reveal the ubiquitination products and substrate input.
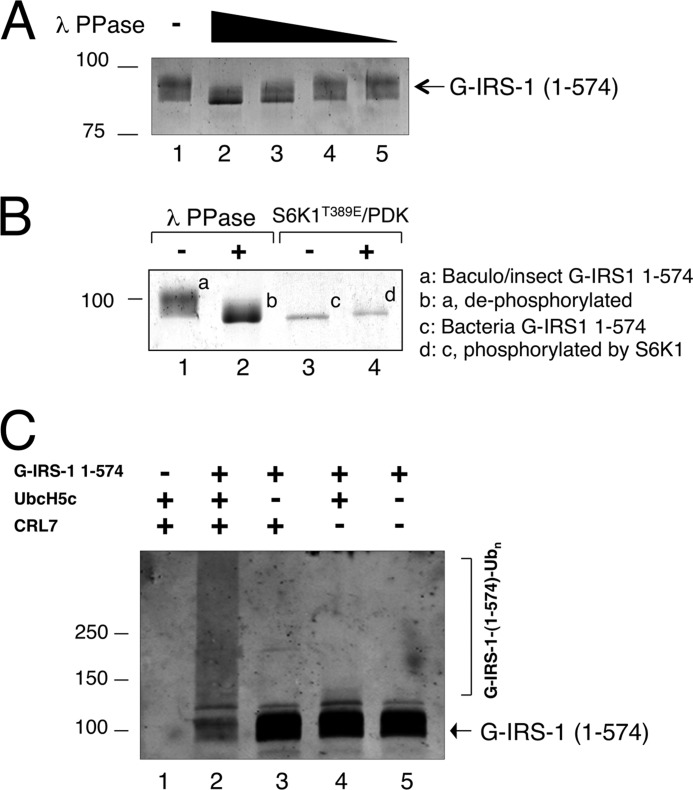
FIGURE 4.
Hyperphosphorylation of IRS1 1–574 activates ubiquitination. A, insect cell-expressed IRS-1 1–574 is hyperphosphorylated. Dephosphorylation reaction was carried out as described under “Experimental Procedures.” The reaction products were analyzed by immunoblot analysis using anti-GST antibody. B, comparison of bacterially or insect cell-expressed IRS1–1-574. Bacterially expressed B-GST-IRS1–1-574, with or without treatment with S6K1, and insect cell-expressed BI-GST-IRS1–1-574, with or without of λ phosphatase, were subjected to SDS-PAGE, followed by Coomassie staining. The various forms of the IRS1 protein are indicated. C, ubiquitination of insect cell-expressed IRS1–1-574. The reaction was carried out as described under “Experimental Procedures.”
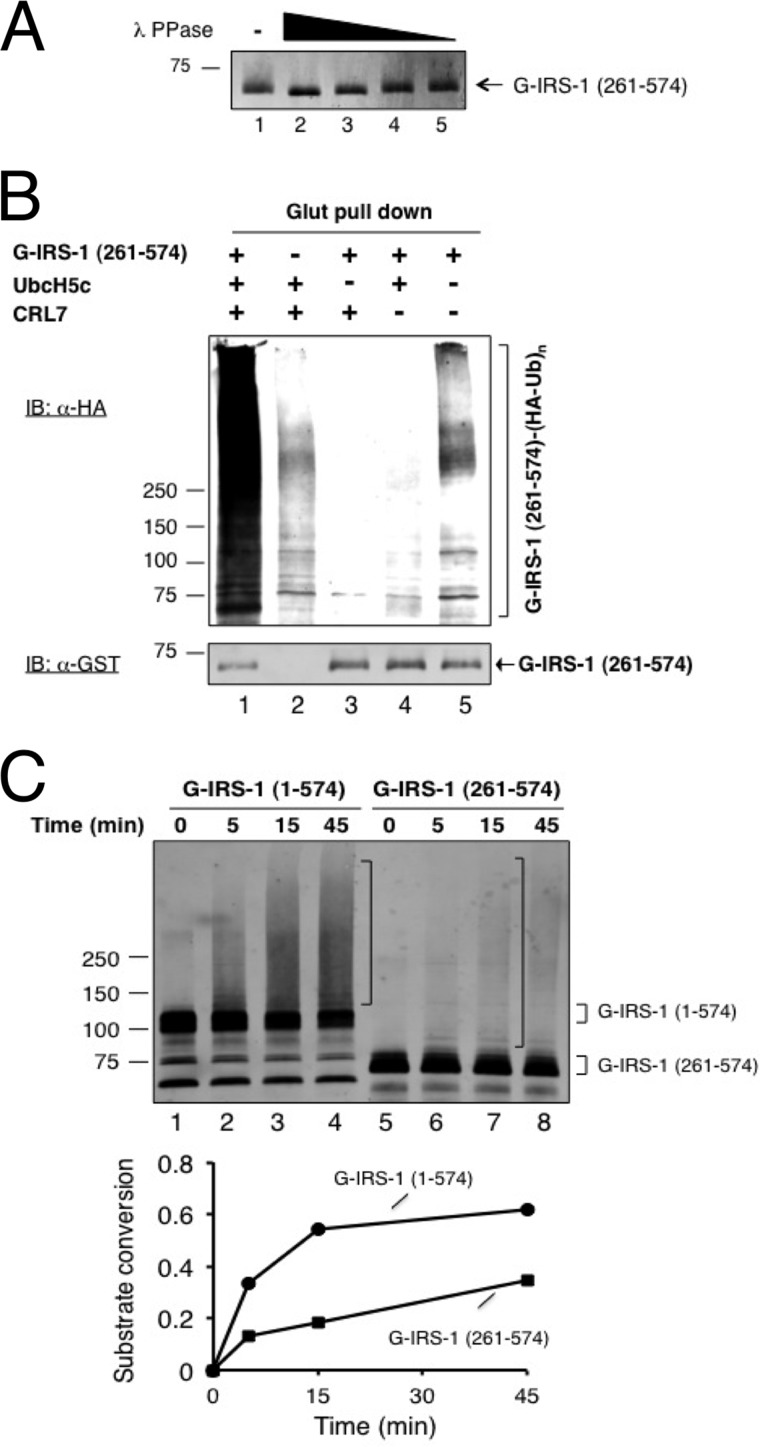
FIGURE 5.
Requirement of the IRS1 N terminus for efficient ubiquitination in vitro. A, insect cell-expressed IRS-1 261–574 is hyperphosphorylated. Dephosphorylation reaction was carried out as described under “Experimental Procedures.” The reaction products were analyzed by immunoblot analysis using anti-GST antibody. B, reconstitution of in vitro polyubiquitination of insect cell expressed IRS-1 261–574 by CRL7. Polyubiquitination of IRS-1 1–574 was reconstituted using glutathione bead enrichment assay as described under “Experimental Procedures.” Immunoblot (IB) analysis with indicated antibodies was performed to reveal the ubiquitination products and substrate input. C, comparison of ubiquitination of insect cell-produced IRS1–1-574 and IRS1–261-574. The time course reaction was carried out as described under “Experimental Procedures.”
For in vitro ubiquitination described in Fig. 3C, B-GST-IRS1–1-574 (5 pmol) was incubated with S6K1T389E/PDK (0.3 μg) at 37 °C for 45 min in a reaction mixture (20 μl) containing 50 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 0.5 mm DTT, 2 mm ATP, 2 mm NaF, and 10 nm okadaic acid. CRL7 (0.13 pmol; shown in Fig. 3A) was added along with PK-Ub (33 μm), E1 (9 nm), and UbcH5c (10 μm), and the final reaction volume was adjusted to 30 μl. Incubation continued at 37 °C for 60 min. To enrich the reaction products, the identical reaction mixture in triplets were combined and absorbed to glutathione-S-Sepharose beads (12 μl). After shaking for 2 h at 4 °C, the unbound proteins were removed by washing. The bound materials were released by boiling the beads for 3 min in SDS loading buffer. The reaction products, separated by 4–20% SDS-PAGE, were analyzed by immunoblot analysis using anti-HA and anti-GST antibodies.
For in vitro ubiquitination described in Figs. 4C and 5C, BI-GST-IRS1–1-574 (1.1 pmol) was incubated in a reaction mixture (20 μl) containing 50 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 0.5 mm DTT, 2 mm ATP, 2 mm NaF, 10 nm okadaic acid, CRL7 (0.2pmol), PK-Ub (50 μm), E1 (13 nm), and UbcH5c (1 μm). Incubation was at 37 °C for times as indicated. The reaction products, separated by 4–20% SDS-PAGE, were analyzed by immunoblot analysis using anti-GST antibodies.
For in vitro ubiquitination described in Fig. 5B, BI-GST-IRS1–261-574 (1.25 pmol) was incubated a reaction mixture (20 μl) containing 50 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 0.5 mm DTT, 2 mm ATP, 2 mm NaF, 10 nm okadaic acid, CRL7 (0.2pmol), PK-Ub (50 μm), E1 (13 nm), and UbcH5c (15 μm). Incubation was at 37 °C for 1 h. To enrich the reaction products, the identical reaction mixture in triplets was combined, adsorbed to glutathione-S-Sepharose beads (12 μl), and analyzed using a protocol identical to that described in Fig. 3C.
Antibodies and Reagents
The following antibodies were purchased: HSP90 (Santa Cruz), V5 (Invitrogen), Myc (Santa Cruz), HA (Roche), IRS-1 (pS307) (Upstate), and GST (Santa Cruz). E1 and MG132 were purchased from Boston Biochem and Sigma, respectively. S6K1T389E/PDK was prepared as described previously (19). PK-Ub (20) and UbcH5c (21) were prepared as described.
RESULTS
In Vivo Analysis of the IRS1 (Fbw8) Degron
Previous studies have implicated several E3 ubiquitin ligases in directing the degradation of IRS1, including SOCS (22), CRL7 (12), Cbl-b (23), and SCFFbxo40 (24). It is thus likely that IRS1 contains multiple distinct sets of degradation determinants (degron) responsible for mediating interactions with their cognate E3s. Elucidation of the specific degron-E3 interactions is key to understanding how discrete cellular signaling pathways impinge on IRS1 homeostasis.
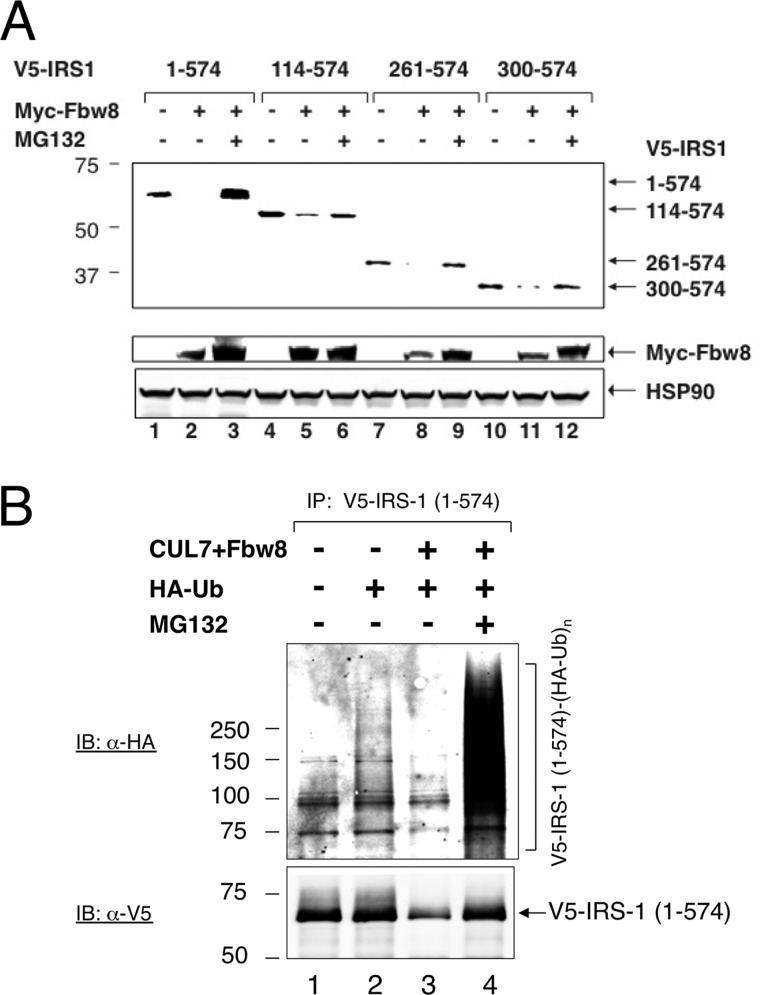
To map the IRS1 (Fbw8) degron, we sought to determine the minimal sequence that confers IRS1 instability in a manner that depends on Fbw8 expression and on proteasomal activity. This strategy has located the IRS1 (Fbw8) degron within the N-terminal half of IRS1, spanning amino acid 1–574, designated as IRS1 1–574 (12). In agreement with previous observations (12), Myc-Fbw8 inhibited the expression of V5-IRS1 1–574, but addition of the proteasome inhibitor MG132 reversed this inhibition (Fig. 1A, lanes 1–3). To further narrow the degron segment, an array of N-terminal truncations were constructed, including V5-tagged IRS1 fragments spanning amino acids 114–574, 261–574, or 300–574. The results showed that similar to V5-IRS1 1–574, V5-IRS1 114–574, 261–574, and 300–574 were all prone to Fbw8- and proteasomal-dependent degradation (Fig. 1A). We have been unable to further narrow the IRS1 (Fbw8) degron by deletion analysis. This is because for reasons unclear to us, further N-terminal truncation of IRS1 300–574 to 392–574 caused instability in a proteasomal independent manner (data not shown).
FIGURE 1.
In vivo analysis of the IRS1 (Fbw8) degron. A, overexpression of Fbw8 induces the degradation of IRS-1 fragments. HEK293 cells were co-transfected with vectors expressing Myc-Fbw8 and V5-IRS-1 truncations as indicated. At 42 h post-transfection, where indicated, the cells were treated with MG132 (10 μm) for 6 h. Extracts (50 μg of protein) were subjected to direct immunoblot (IB) analysis to monitor the changes in the level of V5-IRS-1 and to reveal the presence of Myc-Fbw8. Equal loading was verified by anti-HSP90 immunoblot. B, ubiquitination analysis. HEK293 cells were co-transfected with vectors expressing V5-IRS1–1-574, Myc-Fbw8/FLAG-CUL7, and HA-Ub, singly or in combination. To reveal polyubiquitination, extracts (∼1 mg of protein) from HEK293 cells were immunoprecipitated (IP) with anti-V5 antibodies. The precipitates were separated by 6% SDS-PAGE, followed by immunoblot with anti-HA antibodies.
To evaluate in vivo ubiquitination, V5-IRS1 1–574 was co-expressed with HA-ubiquitin in HEK293 cells. Immunoprecipitation experiments showed that polyubiquitin chains formed on V5-IRS1 1–574 in cells that overexpressed CUL7 and Fbw8 and that has been exposed with proteasome inhibitor MG132 (Fig. 1B). However, despite repeated attempts, we failed to observe ubiquitination of V5-IRS1 261–574 at significant levels (data not shown). Taken together, these results suggest that although IRS1 261–574 and 300–574 contain degron activity, effective ubiquitination/degradation may require the intact IRS1 1–574.
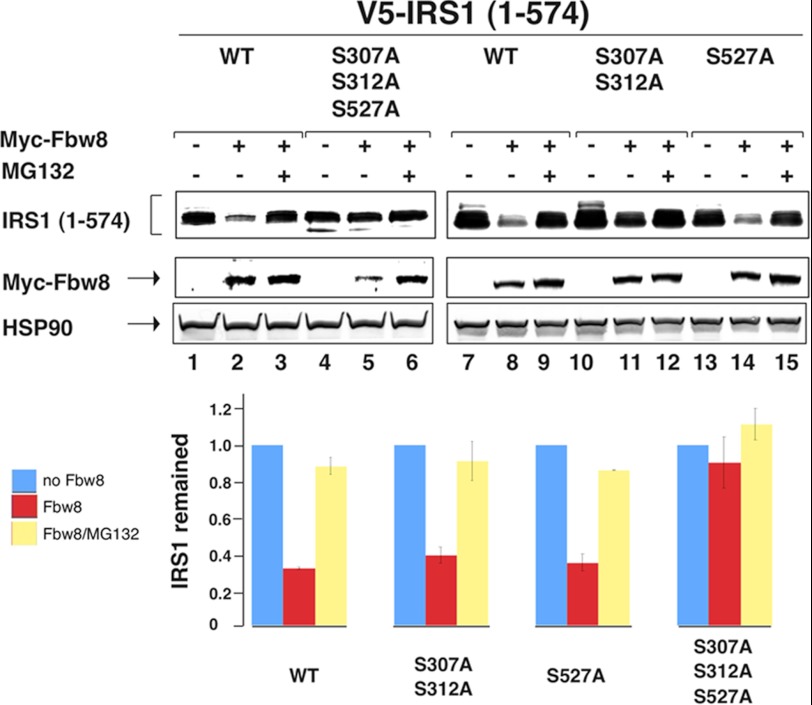
IRS1 1–574 contains a set of well defined mTORC1/S6K1 site at Ser-307, Ser-312, and Ser-527 (25). To assess the role of these residues in Fbw8-dependent degradation of IRS1 1–574, site-directed mutagenesis was carried out to replace these serine residues by alanines. The experiments revealed that the IRS1 mutant bearing serine to alanine substitution at positions 307/312/527 was resistant to degradation (Fig. 2, lanes 4–6). By contrast, either S307A/S312A or S527A was still sensitive to Fbw8-indcued degradation (Fig. 2, lanes 7–15). Thus, the Fbw8-dependent degradation of IRS1 1–574 requires residues Ser-307/Ser-312 and Ser-527.
FIGURE 2.
IRS1 S307/S312/S527 are required for Fbw8-dependent degradation. HEK293 cells were co-transfected with vectors expressing Myc-Fbw8 and V5-IRS1 1–574 or its various point mutants as indicated. The analysis was carried out as described in Fig. 1. The relative intensity between the wild type and point mutants were quantitated and shown graphically. Note that the graph integrates the results of three independent experiments, with error bars for the calculated standard deviation. p < 0.01 (test).
S6K1 Phosphorylation Activates the Ubiquitination of IRS1 1–574 in Vitro, albeit at Low Levels
Decoding the IRS1 (Fbw8) degron requires reconstitution of the IRS1 ubiquitination by CRL7 in vitro. For this purpose, we constructed a stable HEK293-based cell line constitutively expressing FLAG-CUL7 and Myc-Fbw8. Coomassie stain revealed that the affinity-purified CRL7 contained FLAG-CUL7, Myc-Fbw8, Skp1, and ROC1 (Fig. 3A), in keeping with previous findings (12, 18).
Initially, we sought to determine whether ubiquitination could be reconstituted with bacterially expressed GST-IRS1 1–574, designated as B-GST-IRS1 1–574, which is free of eukaryotic post-translational modification. To assess the effect of S6K1 phosphorylation on ubiquitination, we first tested the phosphorylation of B-GST-IRS1 1–574 by using a constitutively active form of purified S6K1 (19). Fig. 3B showed that incubation of B-GST-IRS1 1–574 with S6K1 converted a majority of substrate into slowing migrating forms, which were reactive to anti-phospho-IRS1-Ser-307 specific antibodies, demonstrating that GST-IRS1 1–574 is phosphorylated extensively by S6K1 at sites including the well characterized Ser-307.
To measure ubiquitination, B-GST-IRS1 1–574, treated with or without S6K1, was incubated with affinity-purified CRL7, UbcH5c as E2, E1, and HA-ubiquitin. To enrich the ubiquitin-modified substrates, the reaction mixture was absorbed to glutathione beads. After washing to remove unbound agents, the bound materials were released by denaturation. Ubiquitinated B-GST-IRS1 1–574 was detected by immunoblot analysis with anti-HA antibody. The reaction with the S6K1-treated B-GST-IRS1 1–574 yielded polyubiquitin chains at levels substantially higher than those observed with the unmodified substrate (Fig. 3C, top panel, compare lanes 1 and 4). The production of polyubiquitinated substrates required CRL7 and UbcH5c (Fig. 3C, top panel, lanes 4–6). Anti-GST immunoblot detected B-GST-IRS1 1–574, with the S6K1-treated forms migrating slower than the untreated species (Fig. 3C, bottom panel). No significant levels of high molecular weight, ubiquitin-modified B-GST-IRS1 1–574 forms were detected by anti-GST immunoblot analysis (data not shown). It should be noted that anti-HA-ubiquitin is much more sensitive than anti-GST immunoblot in detecting ubiquitinated conjugates because of their higher reactivity to the former antibodies. Thus, the fact that ubiquitin-modified B-GST-IRS1 1–574 forms were detected by anti-HA-ubiquitin but not by anti-GST immunoblot indicated relatively poor efficiency in this ubiquitination reaction. In all, these results led to the conclusion that S6K1 stimulates the ubiquitination of IRS1 1–574 by CRL7, albeit at low levels.
The Hyperphosphorylated Forms of IRS1 1–574 Support Efficient Ubiquitination by CRL7 in Vitro
The above study suggested that the S6K1 signal alone might not be sufficient to form a highly active IRS1 (Fbw8) degron. In an attempt to generate hyperphosphorylated forms of IRS1 1–574, we overexpressed GST-IRS1 1–574 using the baculovirus/insect cell system and designated it as BI-GST-IRS1 1–574. To evaluate the phosphorylation status of the purified BI-GST-IRS1 1–574, this protein was subjected to dephosphorylation by λ phosphatase. As shown, the treatment by λ phosphatase resulted in substantial increase of the gel migration mobility of BI-GST-IRS1 1–574 in a dose-dependent manner (Fig. 4A). To gain further insights into the extent of phosphorylation of the different preparations of GST-IRS1 1–574, the gel mobility of BI-GST-IRS1 1–574 and B-GST-IRS1 1–574 in their respective modified or unmodified forms were compared directly (Fig. 4B). Evidently, the λ phosphatase-treated BI-GST-IRS1 1–574 exhibited gel mobility identical to that observed with B-GST-IRS1 1–574 (Fig. 4B, compare lanes 2 and 3). However, BI-GST-IRS1 1–574 migrated substantially slower than the S6K1-treated B-GST-IRS1 1–574 (Fig. 4B, compare lanes 1 and 4), suggesting that BI-GST-IRS1 1–574 is in hyperphosphorylated form resulting from multisite phosphorylation by kinases in addition to S6K1.
To reconstitute ubiquitination, BI-GST-IRS1 1–574 was incubated with CRL7, UbcH5c, E1, and ubiquitin. Ubiquitination was measured by direct anti-GST immunoblot analysis. High molecular weight ubiquitination products formed, coinciding with the reduction of the input substrate by nearly 70% (Fig. 4C, lane 2). The reaction required CRL7 and UbcH5c (Fig. 4C, compare lane 2 and lanes 3 and 4). Thus, in contrast to reactions with S6K1-treated B-GST-IRS1 1–574 (Fig. 3), CRL7 directed the ubiquitination of BI-GST-IRS1 1–574 efficiently. These results are consistent with the hypothesis that efficient ubiquitination of IRS1 by CRL7 requires substrate hyperphosphorylation.
Having established BI-GST-IRS1 1–574 as an efficient substrate for CRL7 in vitro, we sought to determine whether N-terminal truncation of this IRS1 fragment affected ubiquitination efficiency. For this purpose, we chose to analyze IRS1 261–574, which exhibited degradation in response to forced expression of Fbw8 (Fig. 1A), but little ubiquitination in vivo (data not shown). GST-IRS1 261–574 was expressed using the baculovirus/insect system and was designated as BI-GST-IRS1 261–574. When subjected to treatment with λ phosphatase, the gel migration mobility of BI-GST-IRS1 261–574 was increased significantly (Fig. 5A), indicative of a hyperphosphorylated status.
We first used glutathione-matrix enrichment assay to measure the ubiquitination of BI-GST-IRS1 261–574 after incubation with CRL7, UbcH5c, E1, and HA-ubiquitin. Anti-HA immunoblot analysis showed the formation of high molecular weight polyubiquitin chains assembled on BI-GST-IRS1 261–574 in a manner that depended on CRL7 and UbcH5c (Fig. 5B, top panel). Anti-GST immunoblot analysis revealed slight reduction of BI-GST-IRS1 261–574 after reaction (Fig. 5B, bottom panel). Thus, in contrast to reactions with BI-GST-IRS1 1–574 that resulted in ∼70% substrate conversion (Fig. 4B), CRL7 mediated the ubiquitination of BI-GST-IRS1 261–574 at substantially lowered levels.
To further compare the ubiquitination efficiency by BI-GST-IRS1 1–574 and BI-GST-IRS1 261–574, we performed a time course experiment. The results showed that BI-GST-IRS1 1–574 exhibited ubiquitination efficiency that was nearly three times higher than that by BI-GST-IRS1 261–574 (Fig. 5C). Thus, the N-terminal 260 amino acids of IRS1 appear to contribute to maximal ubiquitination efficiency.
DISCUSSION
In the mTOR-centered growth regulatory network, IRS1 is required for transmitting signals from the growth factor insulin or insulin-like growth factor 1 to PI3K/Akt, which leads to the activation of mTORC1 and S6K1 (1, 2). In response to hyperactivated mTORC1/S6K1, the well documented mTORC1/S6K1-IRS1 feedback loop induces IRS1 seryl-phosphorylation that promotes its ubiquitin-dependent proteasomal degradation (4). This negative feedback control restrains PI3K/Akt activity and directly impacts the pathogenesis of human diseases including cancer (8) and type 2 diabetes (7). Our previous work has implicated a role for CRL7 in the mTORC1/S6K1-IRS1 feedback loop by targeting IRS1 for ubiquitin-dependent degradation (12). However, it remains elusive how the mTROC1/S6K1 signals are processed to time the IRS1 degradation.
Fig. 6 provides a summary of the IRS1 molecular determinants responsible for triggering CRL7-mediated ubiquitination and degradation, based on previous (12) and the present study using both cell-based degradation and reconstituted ubiquitination approaches. In keeping with our previous observations, we found that forced expression of Fbw8 induced the ubiquitination and degradation of IRS1 1–574 in cells (Fig. 1). Although further N-terminal truncation of IRS1 1–574 by as many as 300 amino acids still retained degron activity (Fig. 1A), detectable cellular ubiquitination was observed only with IRS1 1–574 (Fig. 1B; data not shown). Thus, effective cellular ubiquitination and degradation may require intact IRS1 1–574. In addition, Fbw8-forced degradation of IRS1 1–574 required the integrity of multiple serine residues including Ser-307/Ser-312 and Ser-527 (Fig. 2).
FIGURE 6.
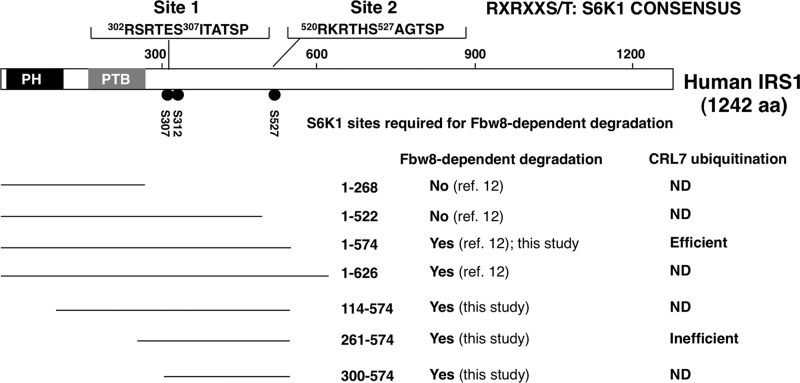
A summary of IRS1 molecular determinants required for CRL7-dependent degradation. The domain structure of human IRS-1 is shown. Both Ser-307/Ser-312 and Ser-527 S6K1 phosphorylation consensus sites are shown, each required for Fbw8-dependent degradation. Seven IRS1 fragments are characterized for Fbw8-dependent degradation, two of which were analyzed for ubiquitination by CRL7 in vitro.
In vitro reconstitution experiments revealed that 1) S6K1 stimulated the ubiquitination of IRS1 1–574 by CRL7, albeit at low levels (Fig. 3). 2) In contrast, the hyperphosphorylated forms of IRS1 1–574 supported efficient ubiquitination by CRL7 (Fig. 4). 3) In comparison with IRS1 1–574, IRS1 261–574 exhibited substantially lowered ubiquitination efficiency despite its hyperphosphorylated status (Fig. 5).
Taken together, the present work helps define several features associated with the IRS1 (Fbw8) degron. First, the IRS1 (Fbw8) degron contains two S6K1 phosphorylation consensus sites centered at Ser-307/Ser-312 and Ser-527 (Fig. 6), each critical for supporting Fbw8-forced degradation (Fig. 2). Second, the S6K1 phospho-IRS1 (Fbw8) degron is partially active, because S6K1 alone was able to activate the ubiquitination of IRS1 1–574 by CRL7, albeit at low levels (Fig. 3). Third, full activity of the IRS1 (Fbw8) degron appears to require hyperphosphorylation by kinases in addition to S6K1 (Fig. 4). Finally, the IRS1 N terminus (amino acids 1–260) is critical for maximal IRS1 (Fbw8) degron activity (Figs. 1 and 5). Of note, our previous work showed that overexpression of Fbw8 did not induce the degradation of IRS1 1–268 (12). Thus, IRS1 1–260 alone is devoid of degron activity but likely plays a role in stabilizing interactions with Fbw8. Additional kinases such as mTORC1 may be required for maximally activating the IRS1 (Fbw8) degron for effective interactions with CRL7. Attempts to examine the effects of mTORC1 on in vitro ubiquitination of IRS1 by CRL7 have been hampered by the lack of recombinant kinase expression/reconstitution systems capable of producing highly active mTORC1.4
Ponyeam and Hagen (26) have recently reported that depletion of CUL7 in cultured cells by siRNA did not alter the protein levels of IRS1. However, it was unclear whether under the conditions used, IRS1 was extensively phosphorylated.
In the mTORC1 signaling, it is essential that the mTORC1/S6K1-IRS1 feedback loop can be fully triggered only when mTORC1/S6K1 reaches toxically high levels. It is possible that IRS1 multisite seryl phosphorylation allows gauging and processing a graded mTORC1/S6K1 signals to mount appropriate responses to control the expression of IRS1, somewhat reminiscent of the degradation of CDK inhibitor Sic1 (27). Recent work by Loog and co-workers has shown the requirement of both Cln2-Cdk1 and Clb5-Cdk1 for Sic1 phosphorylation (28). Cln2-Cdk1 functions as a priming kinase, whereas the action of Clb5-Cdk1 provides positive feedback. It was shown that a subset of Sic1 phospho-sites created by Cln2-Cdk1 served as a docking platform for emerging Clb5-Cdk1, which catalyzed phosphorylation at those Sic1 sites of suboptimal kinetics to Cln2-Cdk1. The synergistic effect between Cln2 and Clb5 has been proposed to greatly amplify the impact of low emerging levels of free Clb5-Cdk1 complexes, creating a hyperphosphorylated form of Sic1 competent for efficient interaction with SCFCdc4, leading to its ubiquitination and degradation that propels the onset of S phase.
A Sic1-like mechanism for activating the IRS1 (Fbw8) degron would enable the discrimination between the signal outputs of mTOC1/S6K1, such that the IRS1 (Fbw8) degron is not maximally activated until sufficient levels of mTOC1/S6K1 activity accumulate. It remains a daunting task to define S6K1/mTORC1 phosphorylation sites on IRS1 and determine how these sites integrate to generate a competent degron that triggers its destruction. Decoding this control mechanism will pave the way toward developing novel therapeutic strategies to modulate mTOR signaling to the benefits of human health.
Acknowledgments
We thank F. Du and I. Tappin for assistance in the construction of baculoviruses expressing human IRS1 N-terminal fragments. We are grateful to J. Nayak for expert technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants CA095634 and GM61051 (to Z.-Q. P.), as well as DK054441 to S.T.
X. Xu and Z.-Q. Pan, unpublished data.
- mTOR
- mechanistic target of rapamycin
- mTORC
- mTOR complex
- S6K1
- S6 kinase 1
- IRS
- insulin receptor substrate
- CRL7
- Cullin-RING E3 ubiquitin ligase 7
- CUL
- cullin
- E1
- ubiquitin-activating enzyme
- E2
- ubiquitin carrier protein
- E3
- ubiquitin-protein isopeptide ligase.
REFERENCES
- 1. Zoncu R., Efeyan A., Sabatini D. M. (2011) mTOR. From growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Laplante M., Sabatini D. M. (2012) mTOR signaling in growth control and disease. Cell 149, 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fisher T. L., White M. F. (2004) Signaling pathways. The benefits of good communication. Curr. Biol. 14, R1005–7 [DOI] [PubMed] [Google Scholar]
- 4. Harrington L. S., Findlay G. M., Lamb R. F. (2005) Restraining PI3K. mTOR signaling goes back to the membrane. Trends Biochem. Sci. 30, 35–42 [DOI] [PubMed] [Google Scholar]
- 5. Shah O. J., Hunter T. (2005) Tuberous sclerosis and insulin resistance. Unlikely bedfellows reveal a TORrid affair. Cell Cycle 4, 46–51 [DOI] [PubMed] [Google Scholar]
- 6. Zhande R., Mitchell J. J., Wu J., Sun X. J. (2002) Molecular mechanism of insulin-induced degradation of insulin receptor substrate 1. Mol. Cell Biol. 22, 1016–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Um S. H., Frigerio F., Watanabe M., Picard F., Joaquin M., Sticker M., Fumagalli S., Allegrini P. R., Kozma S. C., Auwerx J., Thomas G. (2004) Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431, 200–205 [DOI] [PubMed] [Google Scholar]
- 8. Manning B. D., Logsdon M. N., Lipovsky A. I., Abbott D., Kwiatkowski D. J., Cantley L. C. (2005) Feedback inhibition of Akt signaling limits the growth of tumors lacking Tsc2. Genes Dev. 19, 1773–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O'Reilly K. E., Rojo F., She Q. B., Solit D., Mills G. B., Smith D., Lane H., Hofmann F., Hicklin D. J., Ludwig D. L., Baselga J., Rosen N. (2006) mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 66, 1500–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haruta T., Uno T., Kawahara J., Takano A., Egawa K., Sharma P. M., Olefsky J. M., Kobayashi M. (2000) A rapamycin-sensitive pathway down-regulates insulin signaling via phosphorylation and proteasomal degradation of insulin receptor substrate-1. Mol. Endocrinol. 14, 783–794 [DOI] [PubMed] [Google Scholar]
- 11. Greene M. W., Sakaue H., Wang L., Alessi D. R., Roth R. A. (2003) Modulation of insulin-stimulated degradation of human insulin receptor substrate-1 by serine 312 phosphorylation. J. Biol. Chem. 278, 8199–8211 [DOI] [PubMed] [Google Scholar]
- 12. Xu X., Sarikas A., Dias-Santagata D. C., Dolios G., Lafontant P. J., Tsai S. C., Zhu W., Nakajima H., Nakajima H. O., Field L. J., Wang R., Pan Z. Q. (2008) The CUL7 E3 ubiquitin ligase targets insulin receptor substrate 1 for ubiquitin-dependent degradation. Mol. Cell 30, 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sarikas A., Hartmann T., Pan Z. Q. (2011) The cullin protein family. Genome Biology 12, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huber C., Dias-Santagata D., Glaser A., O'Sullivan J., Brauner R., Wu K., Xu X., Pearce K., Wang R., Uzielli M. L.., Dagoneau N., Chemaitilly W., Superti-Furga A., Dos Santos H., Mégarbané A., Morin G., Gillessen-Kaesbach G., Hennekam R., Van der Burgt I., Black G. C., Clayton P. E., Read A., Le Merrer M., Scambler P. J., Munnich A., Pan Z. Q., Winter R., Cormier-Daire V. (2005) Identification of CUL7 mutations in the 3-M syndrome. Nat. Genet. 37, 1119–1124 [DOI] [PubMed] [Google Scholar]
- 15. Huber C., Delezoide A. L., Guimiot F., Baumann C., Malan V., Le Merrer M., Da Silva D. B., Bonneau D., Chatelain P., Chu C., Clark R., Cox H., Edery P., Edouard T., Fano V., Gibson K., Gillessen-Kaesbach G., Giovannucci-Uzielli M. L., Graul-Neumann L. M., van Hagen J. M., van Hest L., Horovitz D., Melki J., Partsch C. J., Plauchu H., Rajab A., Rossi M., Sillence D., Steichen-Gersdorf E., Stewart H., Unger S., Zenker M., Munnich A., Cormier-Daire V. (2009) A large-scale mutation search reveals genetic heterogeneity in 3M syndrome. Eur. J. Hum. Genet. 17, 395–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arai T., Kasper J. S., Skaar J. R., Ali S. H., Takahashi C., DeCaprio J. A. (2003) Targeted disruption of p185/Cul7 gene results in abnormal vascular morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 100, 9855–9860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsunematsu R., Nishiyama M., Kotoshiba S., Saiga T., Kamura T., Nakayama K. I. (2006) Fbxw8 is essential for Cul1-Cul7 complex formation and for placental development. Mol. Cell. Biol. 26, 6157–6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dias D. C., Dolios G., Wang R., Pan Z. Q. (2002) CUL7. A DOC domain-containing cullin selectively binds Skp1·Fbx29 to form an SCF-like complex. Proc. Natl. Acad. Sci. U.S.A. 99, 16601–16606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Keshwani M. M., Ross D. B., Ragan T. J., Harris T. K. (2008) Baculovirus-mediated expression, purification, and characterization of a fully activated catalytic kinase domain construct of the 70kDa 40S ribosomal protein S6 kinase-1 αII isoform (S6K1alphaII). Protein Expr. Purif. 58, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tan P., Fuchs S. Y., Chen A., Wu K., Gomez C., Ronai Z., Pan Z. Q. (1999) Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol. Cell 3, 527–533 [DOI] [PubMed] [Google Scholar]
- 21. Wu K., Fuchs S. Y., Chen A., Tan P., Gomez C., Ronai Z., Pan Z. Q. (2000) The SCFHOS/β-TRCP-ROC1 E3 ubiquitin ligase utilizes two distinct domains within CUL1 for substrate targeting and ubiquitin ligation. Mol. Cell Biol. 20, 1382–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rui L., Yuan M., Frantz D., Shoelson S., White M. F. (2002) SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2. J. Biol. Chem. 277, 42394–42398 [DOI] [PubMed] [Google Scholar]
- 23. Nakao R., Hirasaka K., Goto J., Ishidoh K., Yamada C., Ohno A., Okumura Y., Nonaka I., Yasutomo K., Baldwin K. M., Kominami E., Higashibata A., Nagano K., Tanaka K., Yasui N., Mills E. M., Takeda S., Nikawa T. (2009) Ubiquitin ligase Cbl-b is a negative regulator for insulin-like growth factor 1 signaling during muscle atrophy caused by unloading. Mol. Cell Biol. 29, 4798–4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shi J., Luo L., Eash J., Ibebunjo C., Glass D. J. (2011) The SCF-Fbxo40 complex induces IRS1 ubiquitination in skeletal muscle, limiting IGF1 signaling. Dev. Cell 21, 835–847 [DOI] [PubMed] [Google Scholar]
- 25. Shah O. J., Hunter T. (2006) Turnover of the active fraction of IRS1 involves raptor-mTOR- and S6K1-dependent serine phosphorylation in cell culture models of tuberous sclerosis. Mol. Cell. Biol. 26, 6425–6434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ponyeam W., Hagen T. (2012) Characterization of the Cullin7 E3 ubiquitin ligase. Heterodimerization of cullin substrate receptors as a novel mechanism to regulate cullin E3 ligase activity. Cell Signal. 24, 290–295 [DOI] [PubMed] [Google Scholar]
- 27. Nash P., Tang X., Orlicky S., Chen Q., Gertler F. B., Mendenhall M. D., Sicheri F., Pawson T., Tyers M. (2001) Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414, 514–521 [DOI] [PubMed] [Google Scholar]
- 28. Kõivomägi M., Valk E., Venta R., Iofik A., Lepiku M., Balog E. R., Rubin S. M., Morgan D. O., Loog M. (2011) Cascades of multisite phosphorylation control Sic1 destruction at the onset of S phase. Nature 480, 128–131 [DOI] [PMC free article] [PubMed] [Google Scholar]