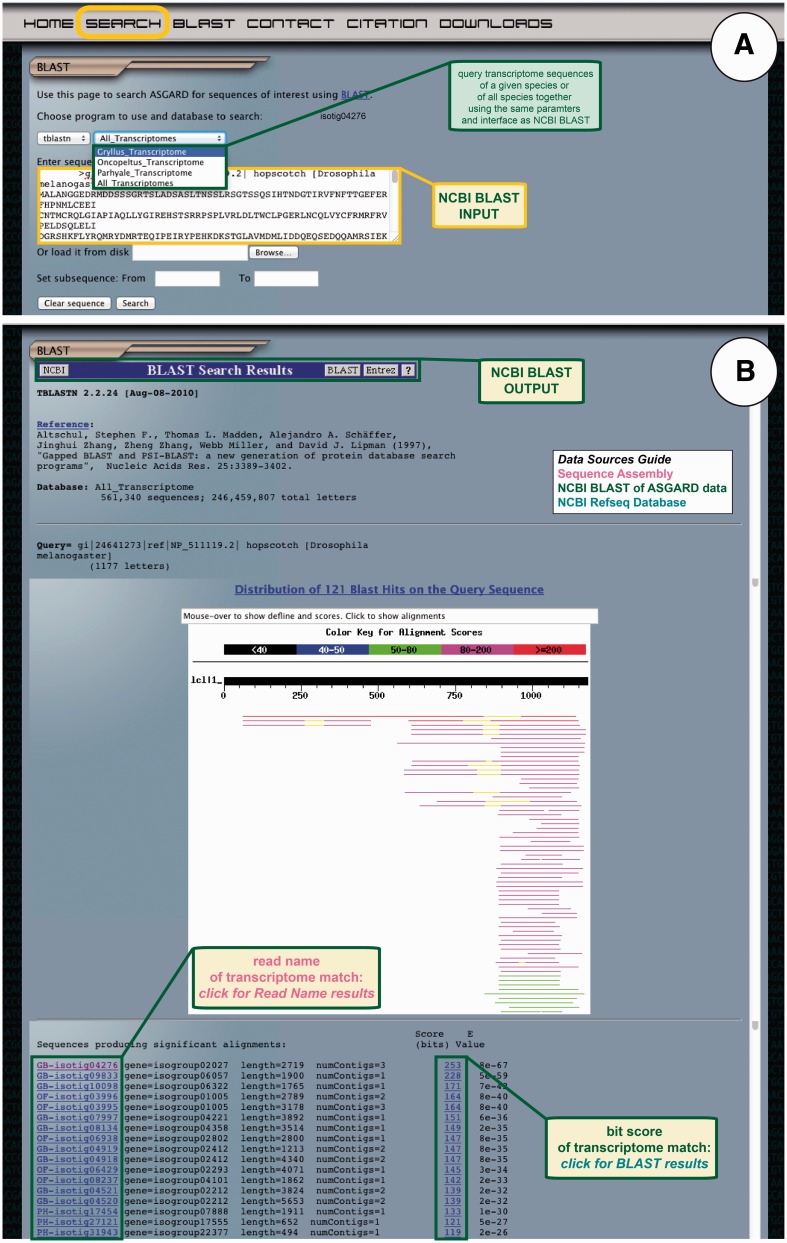
Figure 7.
ASGARD NCBI BLAST search. The BLAST page gives users access to the embedded NCBI BLAST function to query transcriptome sequences with a nucleotide or protein sequence of interest. (A) The input user interface allows users to compare the sequence of any gene of interest to the transcriptome sequences from one or all ASGARD organisms using BLAST. In this example, a user chooses the tBLASTn algorithm to search for G. bimaculatus sequences similar to the D. melanogaster JAK ortholog ‘hopscotch’ by using NCBI accession NP_511119 as a query. (B) The output of this search is transcriptome sequences formatted as for the NCBI BLAST algorithm (97). For each match, the unique identifier links to read name data and the bit score links to the BLAST alignment result. Read names in this output are assigned a prefix identifying the species from which the assembly product derives: GB = Gryllus bimaculatus, OF = Oncopeltus fasciatus, PH = Parhyale hawaiensis. This example shows results of the search for JAK-like G. bimaculatus sequences described in (A).