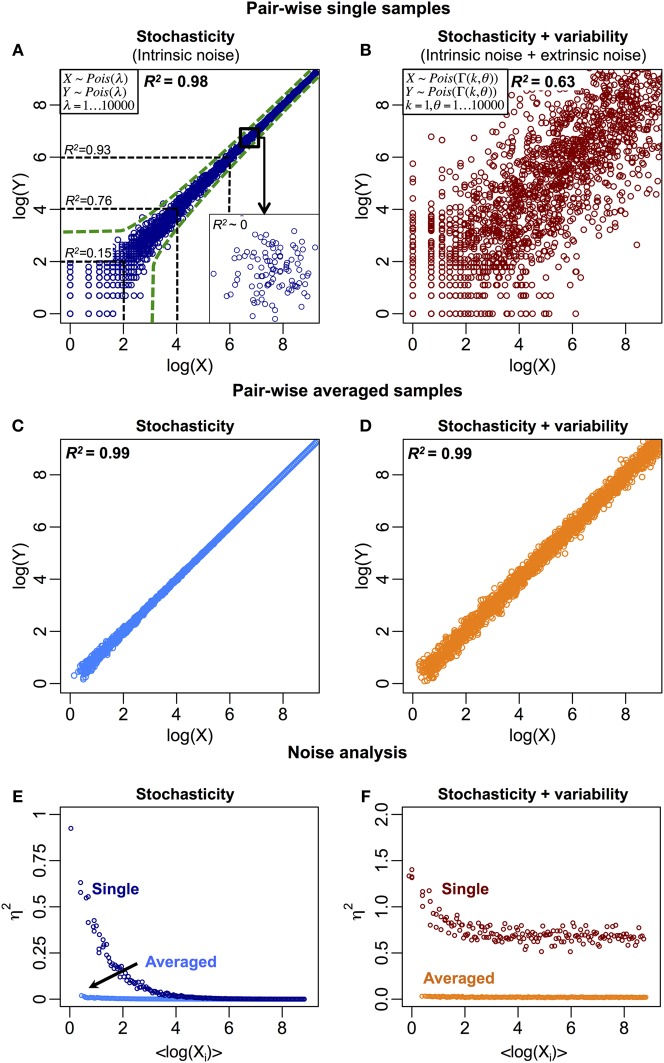
Figure 1.
Biological and non-biological noise reduce the between samples correlation structure. (A) Stochastic fluctuations reduce correlations, especially for low copy number of molecular species (R2 ~0.15 for log(X) < 2). The green dotted lines represent the intrinsic noise region generated by Poisson process (Raj and van Oudenaarden, 2009). Insert: the correlation structure disappears when zooming at smaller or single molecule scale. (B) Stochastic fluctuations (intrinsic) on variable (extrinsic) noise further reduce the overall correlation structure. Variable noise is represented by a Gamma distribution (Taniguchi et al., 2010). R2 is obtained by squaring the Pearson product-moment correlation coefficient, where X = (x1, …, xi, …, xN) and Y = (y1, …, yi, …, yN) are 2 N-dimensional variables, xi and yi are the ith observation (i = 1, …, N) of X and Y respectively. μX and μY are the statistical means of the two variables. (C) Stochastic and (D) total (stochastic and variable) noise reduce when single samples are averaged into population. (E) and (F) show noise, η2 = σ2XY/μ2XY, versus <log(Xi)> for (C) and (D), respectively, where (Jones and Payne, 1997), and the jth element of vectors Xi = (xi, 1, …, xi,j, …, xi,P) and Yi = (yi, 1, …, yi,j, …, yi,P) is the expression of the ith gene in the jth sample for (P = 100) pairs of samples. In (F), at higher expressions for single cells, the remaining noise represents the extrinsic or variable noise. At averaged population scale, this noise is significantly reduced due to the effect of random noise cancellation.