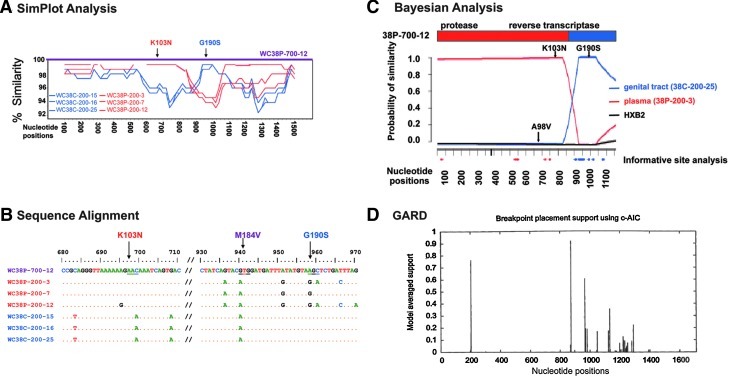
FIG. 2.
Computational analysis of the recombinant HIV-1 genome WC38P-700-12. Viral variants are identified as follows: WC38 denotes Wadsworth Center Subject 38; P or C denotes plasma or CVL as the compartment of origin; numbers that follow denote the month and year the variant was obtained (700 denotes July of 2000); the final number identifies the variant obtained from each compartment on each date. (A) SimPlot compared the similarity of the query sequence, putative recombinant WC38P-700-12, to three representative sequences from the subject's plasma and three from CVL; these representative sequences were obtained in February 2000 and served as most likely parental strains. A graph depicts the degree of similarity of each likely parental strain to the query sequence, with plasma sequences in red and CVL sequences in blue. The SimPlot demonstrates that the portion of the putative recombinant genome WC38P-700-12 encoding the K103N mutation displays ∼99% similarity to the plasma-derived reference sequences; the portion encoding G190S, by contrast, displays an equally strong resemblance to the CVL strains. There is a breakpoint at approximately nucleotide 880 of the RT-protease region. (B) Alignment of the sequence of putative recombinant WC38P-700-12, displayed as the uppermost sequence, with the same three plasma and three CVL-derived most likely sequences displayed in (A). Plasma sequences are depicted in red and CVL sequences in blue. Nucleotides differing from WC38P-700-12 are shown; if identical to that strain, they are depicted as dots. This alignment demonstrates that the sequence of the putative recombinant WC38P-700-12 matches the plasma-derived sequences in the region encoding the K103N mutation and the CVL-derived sequences in the region encoding G190S. Neither plasma nor CVL-derived sequences obtained in February 2000 displayed the M184V mutation seen in July 2000, consistent with the evolution of this mutation between the two time points. (C) Bayesian dual changepoint (BDCP) analysis comparing putative recombinant HIV-1 variant WC38P-700-12 to three other HIV-1 strains: the putative recombinant is compared to two likely parental viral variants derived from the same subject 5 months earlier and the reference strain HXB2. The abscissa represents the alignment of nucleotides of the HIV-1 protease and RT genes; the ordinate displays the probability of similarity between the recombinant strains and the likely parental and reference sequences. The bar diagram at the top represents the protease and RT genes of the recombinant strain, with the breakpoint indicated at approximately nucleotide 880 by a change in color. Sequences from the genital tract (CVL) are shown in blue, those from the plasma are red, and the reference strain HXB2 is black. Resistance mutations displayed by putative parental variants are indicated by arrows. The plasma-derived recombinant variant WC38P-700-12 was compared here to CVL strain WC38C-200-25, plasma strain WC38P-200-3, and HXB2 (posterior probability > 0.99). This figure illustrates that the K103N mutation was contributed by the likely plasma parental strain WC38-200-3 and the G190S mutation by the likely CVL parental strain WC38-C-200-25. Informative site analysis, indicated by red and blue dots along the abscissa, are concordant with the findings of the BDCP analysis. (D) GARD confirms that there is a >90% probability of a recombination breakpoint at approximately the 880 nucleotide position of variant WCP-700-12.