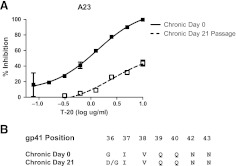
FIG. 5.
Emergence of T-20-resistant virus. (A) T-20 inhibition of chronic phase day 0, nonpassaged (filled squares) versus post-day 21 passaged virus (hollow squares). The x-axis shows the amount of input T-20 in log μg/ml, and the y-axis shows the percent inhibition relative to infection without any inhibitor. Each point represents an average of at least two independent experiments performed in triplicate and is the mean percent inhibition with standard errors of mean. Nonlinear regression was used to estimate a fitted curve in GraphPad Prism5. (B) Predicated amino acids of gp41 envelope positions associated with resistance to T-20. Sequences are shown of bulk PCR envelopes amplified from day 0 and day 21 virus supernatants.