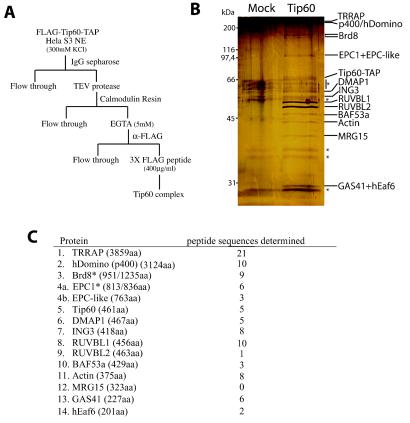
FIG. 2.
Purification of the Tip60-associated proteins identifies the human NuA4 HAT complex. (A) Scheme of the protocol for triple-affinity (using TAP [protein A/calmodulin binding peptide] and FLAG tags) purification of the Tip60-containing complex. (B) Silver-stained gel of mock-treated and triple-affinity-purified material. The Tip60-containing complex was purified from N-terminal FLAG- and C-terminal TAP-tagged Tip60-transduced HeLa cells and then analyzed by silver staining. An extract from nontransduced cells was used as control. Specific bands not present in the control and identified by mass spectrometry and/or Western analysis are labeled on the right. Nonspecific bands are labeled with asterisks. (C) Tip60-associated proteins were identified by tandem mass spectrometry. Tryptic digestion of gel slices from a sample as in panel B were analyzed by tandem mass spectrometry. The proteins identified are listed with the number of different peptide sequences detected for each of them (their lengths in amino acids are indicated in parentheses). Each protein is homologous to a yeast NuA4 subunit, with the exception of Brd8 and RUVBL1/2. MRG15, did not produce peptide hits but was identified by Western analysis. Proteins with asterisks indicate the detection of protein products from splice variants. One mass spectrometry hit was specific for the shorter isoforms of both Brd8 (also known as p120) and EPC1 (also known as EPC2). p400 is named hDomino because of the close homology with mouse and Drosophila Domino proteins (43). Identified human protein FLJ11730 is named hEaf6 because of the homology with the yeast NuA4 subunit Eaf6. The accession number for the EPC1 paralog named EPC-like is NP_056445.