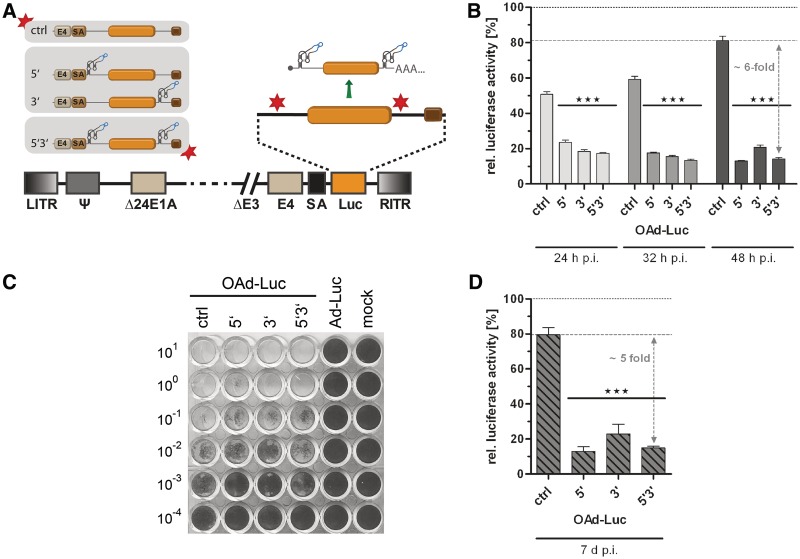
Figure 4.
Application of the aptazyme for regulation of late transgene expression by oncolytic adenoviruses (OAds). (A) Schematic outline of replication-competent OAds. The firefly luciferase gene (orange box) with SV40 polyadenylation signal (brown box) was inserted with an upstream splice acceptor site (SA) into the OAdV genome, facilitating transgene co-expression at the late phase of virus replication. E1AΔ24, E1A gene with 24 bp deletion for tumor-selectivity. E4, E4 region. Other abbreviations as in Figure 3. Nomenclature for aptazyme and control constructs is as in Figure 2. (B) A549 cells were infected with OAds at 5 TCID50/cell and cultured in the presence or absence of 3 mM theophylline. Cells were harvested at 24 h, 32 h and 48 h post infection. Shown are relative luciferase activities for OAds in the presence of theophylline in % of expression levels in the absence of theophylline (set as 100% for each time point, dotted line) of a representative experiment. Dashed line, reference gene activity level for calculation of aptazyme performance. Columns/symbols show mean values of triplicate infections, error bars SD. Significance for both theophylline-dependent down-regulation of gene expression for individual constructs and relative reporter gene expression in comparison with ctrl is indicated with ***(P < 0.001). (C) Spread-dependent cytotoxicity of OAds in A549 cells as determined by crystal violet staining of surviving cells 20 d post infection. Numbers are viral titers in TCID50/cell used for infection. Ad-Luc, replication-deficient virus (AdV ctrl, see Figure 3). Mock, mock-infected cells. (D) A549 cells were infected using 0.05 TCID50/cell. Five days post infection, media was removed, and cells were cultured for 2 more days either in the absence or presence of 3 mM theophylline. Data presentation is as in panel B.