Figure 3.
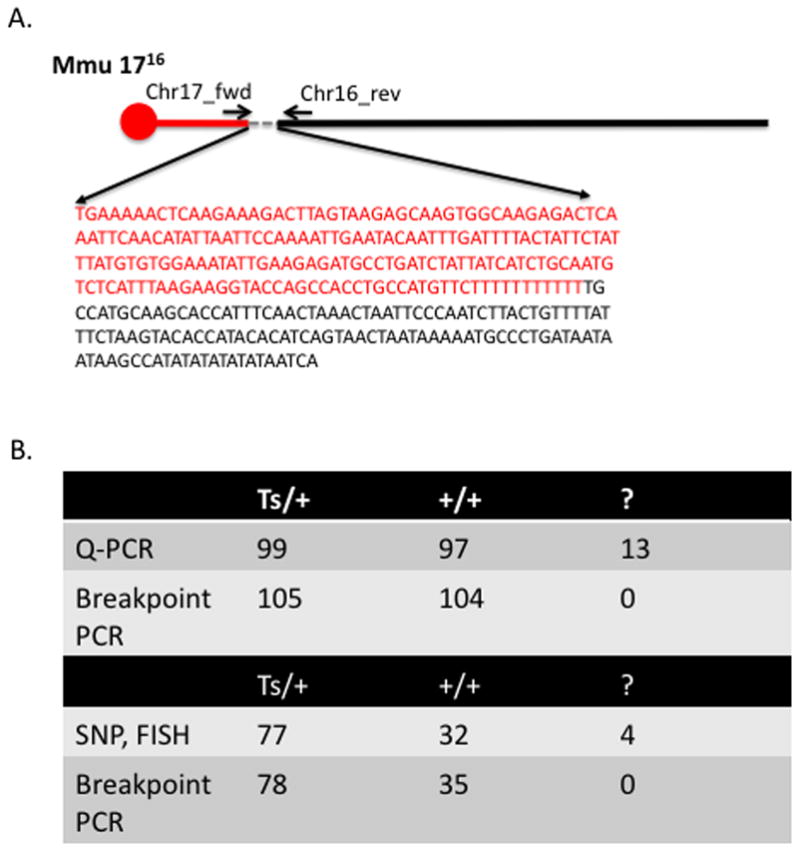
Amplification and sequencing of the 1716 junction. PCR and Sanger sequencing of the junction confirmed the precise sequence of the breakpoint as predicted by de novo assembly of reads spanning the junction (A). PCR amplification of the junction in 209 previously genotyped samples were completely concordant with samples that could be confidently scored as trisomy (Ts/+) or euploid (+/+) by quantitative PCR (Q-PCR) (Liu et al. 2003). Moreover, 13 genotypes (‘?’) that could not be scored due to inconclusive Q-PCR results were successfully scored as either trisomy or euploid based on amplification of the junction (“breakpoint PCR”). Similarly, PCR amplification of the junction in an additional set of 113 previously genotyped samples were also completely concordant with samples that could be confidently scored as trisomy (Ts/+) or euploid (+/+) by SNP prescreen (Lorenzi et al.) and DNA FISH (Moore et al. 1999). In addition, 4 genotypes (‘?’) that could not be confidently genotyped via SNP prescreen or for which SNP prescreen results and DNA FISH were conflicting, could be confidently scored as either trisomy or euploid by PCR amplification of the 1716 junction (B).
