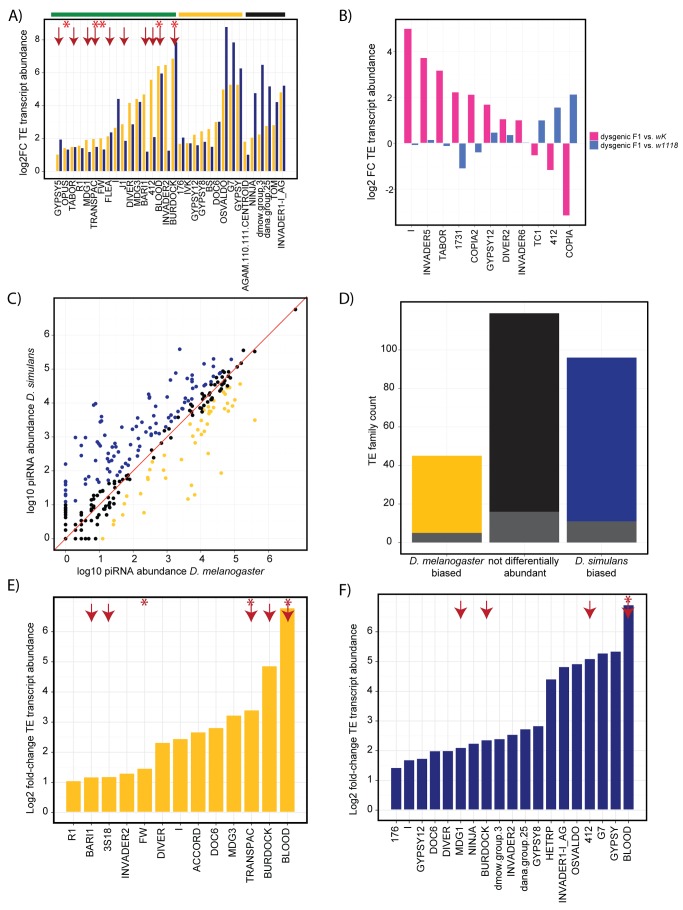
Figure 1. The pattern of TE derepression in interspecific hybrid ovaries does not correlate with interspecific differences in piRNA abundance and does not match expectations from models of intraspecific hybrid dysgenesis.
(A) Widespread misexpression of TEs in interspecific hybrids. Log2 transformed ratio of TE transcripts exhibiting ≥2-fold increased transcript abundance in interspecific F1 hybrids relative to both D. melanogaster (yellow) and D. simulans (blue). Candidate horizontally transferred TE families that are derepressed in interspecific hybrids are highlighted by a red arrow [10] and/or a red asterisk [12]. TE families represented by full-length copies in both genomes, the D. melanogaster genome only, or neither genome are indicated under the green, yellow, and black bars, respectively [10],[49]. (B) No TEs in dysgenic intraspecific hybrids exhibit ≥2-fold increased transcript abundance relative to both parental strains. Log2 transformed fold-change in TE transcript abundance in F1 dysgenic hybrids relative to D. melanogaster wK (pink) and w1118 (blue) parental strains. (C) Many TE classes have differential abundance of their corresponding piRNAs between D. melanogaster and D. simulans. D. melanogaster biased TEs (≥2-fold higher abundance) are yellow, and D. simulans biased TEs are shaded blue. Red line indicates equivalent expression values in both species. (D) TE derepression in interspecific hybrid ovaries does not correlate with interspecific differences in piRNA abundance. Each bar represents the total number of TE classes with the pattern of interspecific differential abundance for piRNAs from (C), while grey shading indicates the proportion that are misregulated in interspecific hybrids from (A). The proportion of misregulated TEs is not significantly different among the three classes (X2 = 2.08, df = 2, p = 0.35). (E and F) Species-specific TE transcripts are equivalently derepressed from the maternal and paternal genomes in interspecific hybrids. Log2 transformed ratio of TE transcripts exhibiting ≥2-fold increased abundance in hybrids relative to D. melanogaster (yellow) or D. simulans (blue) when considering reads that map exclusively to the D. melanogaster or D. simulans genomes. Red arrows and asterisks as in (A).