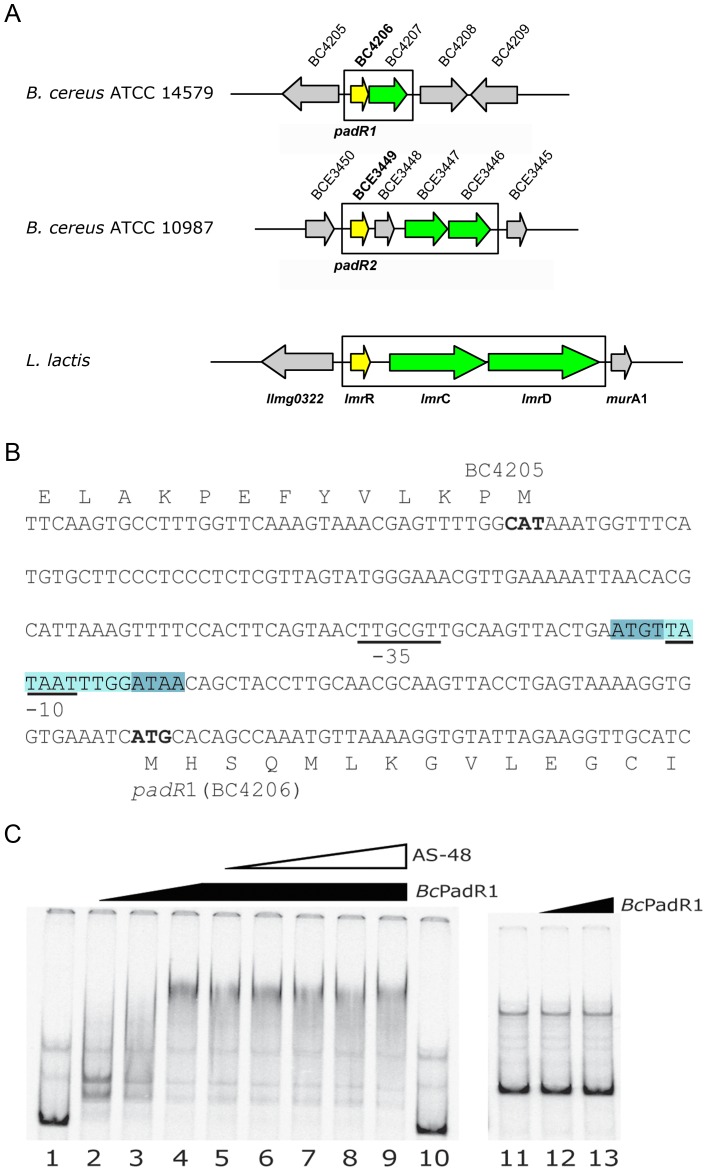
Figure 1. Genomic neighborhood of the bcPadR1 and bcPadr2 encoding genes and analysis of promoter binding.
(A) Organization of the BC4206(bcPadR1)-BC4207 operon of B. cereus ATCC 14579, the putative BCE3449(bcPadR2)-BCE3448-BCE3447-BCE3446 regulon of B. cereus ATCC 10987 and the lmrR-lmrCD regulon of L. lactis. The relative scale of the genes and intergenic regions is proportional to nucleotide length. Boxes indicate the operon/regulon boundaries. The PadR-encoding genes are colored in yellow, while their (putative) target genes, encoding resistance-associated membrane proteins, are in green. (B) The sequence of the intergenic region between BC4205 and BC4206 that is used for the EMSA experiments. Putative -35 and -10 promoter sequences of BC4206 are underlined, the start codons of BC4205 and BC4206 are highlighted in bold, and the amino acids of the coded proteins are indicated above the DNA sequence. The putative binding site of bcPadR1 (homologous to the canonical ATGT/ACAT inverted sequence motif) is highlighted in blue. (C) EMSA experiments with bcPadR1. DNA fragments encompassing the promoter regions of BC4206 (lanes 1–10) and BC4029 (lanes 11–13, negative control) were prepared by PCR and end labeled with 33P. DNA binding was assayed as described in the Materials and Methods. Lanes 1, 10 and 11 contain the free DNA probe. Samples run on lanes 2 and 12 contain 0.5 µM; lane 3 1 µM; lanes 4–9 and 13 2 µM of purified bcPadR1 protein. Lanes 5 to 9 contains samples including increasing concentrations of AS-48 from 0.5 pM to 0.69 µM.