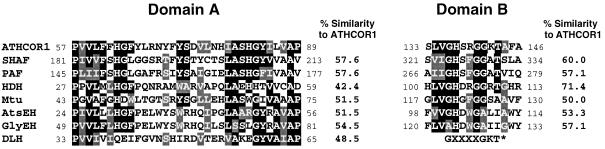
Figure 3.
Protein alignments between regions of the translated peptide from ATHCOR1 with two conserved domains (A and B) found in different hydrolases. SHAF, Similar to human-activating factor acetylhydrolase from C. elegans, U64598; PAF, platelet-activating factor acetylhydrolase from Cavia porcellus, JC5021; HDH, haloacetate dehalogenase from Moraxella sp., A44856; Mtu, unknown Mycobacterium tuberculosis Z95389; AtsEH, Arabidopsis epoxide hydrolase, D16628; GlyEH, soybean epoxide hydrolase, D63781; and DLH, dienelactone hydrolase from Synechocystis sp. dienelactone hydrolase, D90904. Black boxes indicate conserved amino acids at the minimum of 50%. Gray boxes indicate changes by similar residues. The percentage of similarity between ATHCOR1 and each of the sequences is presented. The consensus for the putative ATP-/GTP-binding site described in the literature (Saraste et al., 1990) is indicated by an asterisk. Sequences were aligned by the CLUSTAL W program (Thompson et al., 1994) and shaded by the BOXSHADE program (used at the Bioinformatics Group WWW site at the Swiss Institute for Experimental Cancer Research, Lousanne, Switzerland).