Abstract
The lysine deacetylase family of enzymes (HDACs) was first demonstrated to catalyze deacetylation of acetyllysine residues on histones. In subsequent years, HDACs have been shown to recognize a large pool of acetylated non-histone proteins as substrates. Recently, thousands of acetylated proteins have been discovered, yet in most cases, the HDAC that catalyzes deacetylation in vivo has not been identified. This gap has created the need for better in vivo, in vitro, and in silico approaches for determining HDAC substrates. While HDAC8 is the best kinetically and structurally characterized HDAC, few efficient substrates have yet been substantiated in vivo. In this review we delineate factors that may be important for determining HDAC8 substrate recognition and catalytic activity, including structure, complex formation, and post-translational modifications. This summary provides insight into the challenges of identifying in vivo substrates for HDAC8, and provides a good vantage point for understanding the variables important for predicting HDAC substrate recognition.
Introduction
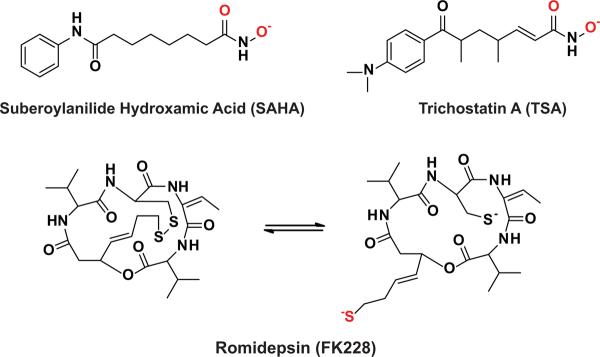
Acetylation of lysine side chains in proteins is a reversible post-translational modification that occurs in a wide range of organisms1. This modification affects the properties of proteins, including protein-protein association, protein-DNA interactions, and protein stability2. Initially, acetylation gained recognition as a post-translational modification to histones. Acetylation of histones can regulate the accessibility of DNA to cellular machinery and thus change the protein expression profiles of cells3. Due to the effect of acetylation on the proteome, it is not surprising that many diseases have been associated with the aberrant acetylation of histones4. In the last twelve years the paradigm for protein acetylation has changed drastically, moving from a histone centric model to a proteome centric model. This change in mindset has resulted from the identification of acetylated lysine side chains that affect the function of numerous non-histone proteins5–6. Currently, over 3,600 acetylation sites have been discovered in mammalian proteins7 and these protein are important in many cellular processes, including gluconeogenesis and DNA damage repair5–6. Regulation of the acetylation state of proteins is important as aberrant acetylation of both histone and non-histone proteins can contribute to the development of many disease states8–10. As proof of this, two broad spectrum HDAC inhibitors (suberoylanilide hydroxamic acid (SAHA) and Romidepsin) have been approved by the FDA and are currently on the market for the treatment of T-cell Lymphomas [Figure 1]11.
Figure 1.
Widely used HDAC inhibitors. SAHA and Romidepsin have been approved by the FDA for use as second line treatments for T-cell lymphomas. TSA is an inhibitor that has been used widely in in vitro and in vivo studies but is not being tested in drug trials105. All three inhibitors are competitive with substrates by occupying the substrate binding channel and coordinating the active site metal ion. The atoms colored red interact with the active site metal ion54–55,63–67.
HDAC isozymes can be grouped into four classes based on their phylogenetic similarity12. Class I (HDAC1, 2, 3, and 8), class IIa (HDAC4, 5, 7, and 9), class IIb (HDAC6 and 10), and class IV (HDAC11) enzymes catalyze deacetylation using a metal dependent mechanism12–13, while class III (Sirt1-7) enzymes use an NAD+ cofactor to perform deacetylation14–15. Due to the abundance and importance of HDAC substrates, one of the foremost questions in the field is the determination of the substrate specificity of HDACs. This area of research seeks to identify which of the 18 deacetylases catalyzes deacetylation of each of the >3,600 mammalian acetylation sites. Adding to the complexity of this problem is the possibility that cellular regulation may alter both the catalytic activity and the substrate specificity of HDACs. Illuminating the substrate selectivity and regulation of HDACs should shed light on the mechanism and treatment of acetylation-related diseases.
Mechanistically and structurally, HDAC8 is the best studied of the HDAC homologues. Furthermore, HDAC8 is proposed to recognize a number of non-histone substrates16–18 and is therefore a good model for developing techniques to unravel HDAC substrate specificity. In this review we discuss the current view of HDAC8 regulation, and compare HDAC8 to other promiscuous enzymes to identify factors that determine substrate specificity.
Known HDAC8 substrates
HDAC8 was initially discovered in 2000 and was shown to catalyze in vitro deacetylation of a number of acetylated histone variants19–21. These substrates included full-length H2A/H2B, H3, and H4 histones, acetylated at non-specific lysines19–20. Concurrent studies showed that peptide sequences corresponding to the H4 histone tail with an acetylated lysine at position sixteen (K(Ac)16) were also in vitro substrates20–21. In subsequent years, several studies have used the H4 histone tail sequence as a peptide template to investigate the amino acid sequence preference of HDAC8 (discussed below)22–24. Recently, HDAC8 was demonstrated to catalyze in vitro deacetylation of the K(Ac)20 site on the H4 histone tail. However, HDAC8-catalyzed deacetylation of the K(Ac)20 peptide is much slower than deacetylation of K(Ac)16 peptides25, suggesting that another HDAC isozyme may catalyze this reaction in vivo. Despite these findings, the role of HDAC8 in catalyzing deacetylation of specific sites in histones in vivo remains unclear.
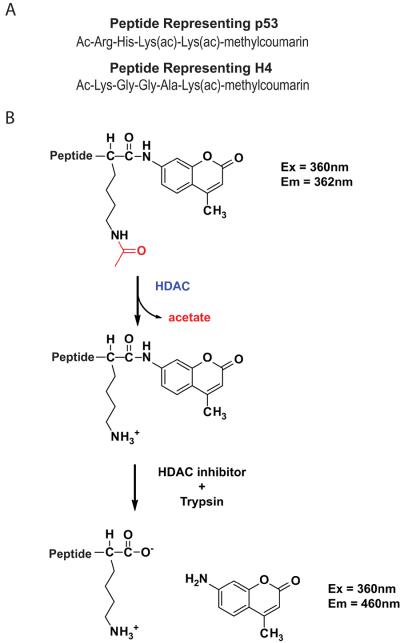
Shortly after HDAC8 was identified, the first non-histone acetylated proteins were reported26–27 which inspired researchers to hunt for other possible HDAC substrates. The search for new HDAC8 substrates was further spurred by the finding that this enzyme is present in the cytoplasm of smooth muscle cells28–29, causing evaluation of non-nuclear substrates. In fact, HDAC8 catalyzes deacetylation of a peptide corresponding to the C-terminal end of the p53 transcription factor [Figure 2a] faster than the K(ac)16 H4 histone peptide [Biomol, unpublished]. HDAC8 catalyzes deacetylation of coumarin derivatives of the acetylated p53 and H4 peptides with kcat/KM values of 7,500 M−1s−1 18 and 2,800 M−1s−1, respectively30. Since the kcat/KM parameter reflects the relative reactivity of an enzyme with different substrates31, these values suggest that HDAC8 has a modest preference for catalyzing deacetylation of p53 over the H4 histone. It is important to note that these kcat/KM values for HDAC8 were measured using the commercially available Fluor-de-lys assay (Biomol). This assay uses peptide substrates containing a methylcoumarin fluorophore conjugated to the C-terminal side of the acetyllysine residue. After deacetylation, digestion by trypsin cleaves the coumarin fluorophore, causing an increase in fluorescence at 460 nm; deacetylation is measured from an increase in the fluorescence signal32 [Figure 2b]. While this assay has been a valuable tool for studying histone deacetylases, the methylcoumarin fluorophore increases the reactivity with HDAC833. Therefore, deacetylation of the nonlabeled acetylated p53 and H4 histone peptides catalyzed by HDAC8 may be slower than reported using this assay. Furthermore, the coumarin substrates may not reliably reflect HDAC substrate specificity in the context of full-length proteins.
Figure 2.
The Fluor de lys assay [Biomol]32. A. The sequence of two HDAC8 substrates used for the Fluor de lys assay. B. Schematic of the Fluor de lys assay, including the wavelengths used to measure the methylcoumarin fluorophores.
The steady state kinetic parameters for catalysis of the deacetylation of peptides can provide insight into both the kinetic mechanism and the in vivo reactivity of these substrates. HDAC8-catalyzed deacetylation of the p53 and H4 coumarin peptides has a low value of kcat/KM (103 – 104 M−1s−1) in comparison to enzymes that function near diffusion-controlled limits (106 – 108 M−1s−1) and a high value for KM (320 μM, H4 peptide)30 compared to other HDAC isozymes (~30 μM)34. These data suggest a simple Michealis-Menten kinetic model whereby substrate binding and dissociation is rapid, and is followed by rate-limiting deacetylation. This conclusion is bolstered by the observed enhancement of the kcat value for deacetylation of peptides labeled with a more reactive trifluoroacetyl group35–36. Therefore, substrate specificity is determined by both the affinity of HDAC8 for a peptide substrate and the reactivity of the enzyme-substrate complex. Assuming that the kinetic constants for deacetylation of these peptides mimic the full-length proteins, the low kcat/KM and high KM values for the H4 and p53 peptides compared to reactivity with other isozymes18,30,34 suggest that HDAC8 may not catalyze deacetylation of these sites in vivo. However, it is possible that natural, full length, substrates may be better optimized for efficient deacetylation to allow for regulation of these posttranstlational modifications. Cellular data implicating HDAC8-catalyzed deacetylation of H4 and p53 in vivo is also sparse. In addition to these proposed substrates, in vitro kinetic studies combined with cellular assays have yielded several promising candidates for in vivo HDAC8 substrates (discussed further below).
There are a number of factors that must be taken into account when parsing whether substrates are acted upon by a given enzyme in vivo. HDAC selectivity is minimally described by the relative values of kcat/KM for deacetylation, the relative concentrations of the HDAC isozymes, and the concentrations of competing substrates. Relative kcat/KM values indicate the substrate preference of an enzyme when discriminating amongst multiple substrates31. The majority of enzymes have kcat/KM values of 105–106 M−1s−1 37. These values are generally slower than the diffusion controlled rate constants for substrate binding, which can be as high as 107–108 M−1s−1 31. Consistent with this, the kcat/KM values for the HDAC8 homolog HDAC1 for deacetylation of the peptide,Ac-Gly-Ala-Lys-AMC and, the homologous enzyme, arginase I are on the order of 105 M−1s−1 34,38, suggesting that similar values should be achievable for efficient HDAC8 substrates. One caveat to making conclusions from kinetic parameters measured in in vitro experiments is that some enzymes require an activator for optimal activity. Since many HDAC isozymes associate with large protein complexes in vivo, it is possible that other proteins in the complex could activate the catalytic activity or enhance the substrate affinity to increase the value of kcat/KM in the cell.
Candidate non-histone HDAC8 Substrates
One promising HDAC8 substrate is the Estrogen-Related Receptor α (ERRα). This orphan receptor is expressed in a number of organs, including the heart, kidney, and muscle, where it controls processes that are essential for maintaining energy homeostasis39. ERRα can be acetylated at four lysines, where these post-translational modifications inhibit DNA binding17. A role for HDAC8 in catalyzing the deacetylation of ERRα was suggested by the demonstration that the acetylation state of ERRα was altered by simultaneous incubation with HDAC8, the histone acetyltransferase PCAF, and 14C-acetyl-CoA17. Furthermore, incubation of purified acetylated-ERRα with HDAC8 enhances the affinity of ERRα for DNA, which is consistent with HDAC8-catalyzed deacetylation of ERRα. One caveat to these experiments is that this assay included metal chelators and low salt, conditions where HDAC8 has limited catalytic activity18,40. An alternative explanation of these data is that HDAC8 binds to ERRα to increase the DNA affinity and decrease acetylation catalyzed by PCAF. However, addition of the non-homologous deacetylase, Sirt1, to these in vitro assays also decreases acetylation of ERRα, suggesting that both enzymes recognize ERRα as a deacetylase substrate. Finally, RNAi-dependent decreases in cellular HDAC8 or Sirt1 levels are accompanied by increases in ERRα acetylation in vivo17. Taken together, these results suggest that HDAC8 catalyzes deacetylation of ERRα in vivo. Consistent with this, the acetylation site (K129(ac)) in ERRα has Arg in the −1 position (the amino acid on the N-terminal side of the acetyllysine), and RK(ac) motifs have been demonstrated to be favorable for HDAC8 catalysis33. Additional analysis, such as directly measuring ERRα acetylation patterns using mass spectrometry in the presence and absence of HDAC inhibitors would further validate ERRα as an in vivo substrate of HDAC8.
Another proposed HDAC8 substrate is the aberrant inv(16) fusion protein found in a significant portion of patients with acute myeloid leukemia41. This fusion protein combines the N-terminus of the transcription factor domain core binding factor β, with the C-terminus of the smooth muscle myosin heavy chain42. In COS7 cells, co-immunoprecipitation experiments demonstrated that overexpressed HDAC8 associates with inv(16)16. Furthermore, HDAC8 co-localizes and immunoprecipitates with smooth muscle myosin heavy chain43 suggesting that HDAC8 may interact with this domain within the inv(16) fusion protein. Other HDAC isozymes do not immunoprecipitate with inv(16) under similar conditions, which suggests that HDAC8 may be the main HDAC that interacts with inv(16) in vivo. The addition of the HDAC inhibitor TSA inhibits the transcriptional repression activity of inv(16)16, suggesting that HDAC8 activity is important for inv(16) regulation. An alternative explanation of these data is that inv(16) is a binding partner with HDAC8 rather than a substrate, as HDAC inhibitors have been shown disrupt the association of HDACs with non-substrate binding partners44. The acetylation site in the core binding factor β is RSK(ac)FE5. Peptide library studies have demonstrated that Phe in the +1 position is favorable for HDAC8 catalysis23,33 although Ser at the −1 position attenuates reactivity33. While the core binding factor β is acetylated in vivo5, there is not yet direct evidence that inv(16) is acetylated45. Taken together, these data indicate that inv(16) is either an HDAC8 substrate or forms a functionally important complex with HDAC8.
A third potential in vivo HDAC8 substrate is the transcription factor CREB. Acetylation at three CREB sites (Lys91, Lys96, and Lys136) helps to activate this protein46. HDAC8 and CREB overexpressed in HEK293 cells co-immunoprecipitate, demonstrating that these two proteins associate. When HDAC8 is overexpressed in cells, phosphorylation of CREB decreases, which in turn inhibits CREB transcriptional activation47. Likewise, treatment of cells with the HDAC inhibitor TSA increases CREB phosphorylation levels48 suggesting that HDAC8 activity is important for CREB phosphorylation. However, the addition of a broad range HDAC inhibitor (such as TSA) decreases the activity of all metal-dependent HDACs. As HDAC overexpression can affect a number of targets within the cell, this inhibition may indirectly affect CREB phosphorylation. Furthermore, pulldown experiments demonstrate that CREB can interact with a number of HDAC isozymes47, complicating identification of CREB as an HDAC8 substrate in vivo. Due to the high amino acid identity between class I HDACs (>30%)49, overexpression and pulldown experiments may not yield results that are representative of in vivo situations. Therefore, these experiments suggest, but do not confirm, a direct connection between HDAC8 deacetylase activity, the phosphorylation status of CREB, and the regulation of CREB activation. Alternatively, HDACs may function as protein scaffolds to mediate the inhibitory interaction between CREB and PP1 phosphatase47,50–52, leading to a decrease in CREB phosphorylation and activity.
The current cellular methods for identifying substrates of HDAC isozymes in vivo have limitations. Since HDAC selectivity depends on the relative concentrations of the HDAC isozymes and the concentrations of all of the acetylated lysine substrates, overexpression of HDAC and/or HDAC substrates can alter the normal pattern of deacetylase activity. Therefore, experiments using overexpressed proteins can suggest that a particular interaction occurs in vivo, but does not prove that this contact occurs under physiological conditions. Native pulldown experiments, which should be more representative of physiological conditions, have thus far not been successfully used to confirm the identity of HDAC8 substrates. It is possible that the HDAC-substrate interactions may be transient and/or weak and thus are not maintained through the multiple washes in pulldown experiments. Therefore alternate techniques, such as crosslinking, may be necessary to increase the lifetime of an HDAC-substrate complex to allow detection. Additionally, observation of enhanced acetylation after deletion or knockdown of a given isozyme does not prove that an HDAC isozyme directly catalyzes deacetylation of that site. Therefore alternative methodologies need to be explored to enhance the identification of additional HDAC8 substrates.
HDAC8 complex formation
Recombinantly purified HDAC8 catalyzes deacetylation and displays substrate selectivity in the absence of additional protein cofactors18–20,22–24,33–34,36,40,53–55, suggesting that HDAC8 can catalyze deacetylation in vivo in the absence of a protein complex. In contrast, the other class I HDACs, HDAC1, 2, and 3, are observed in complexes in the cell and their substrate specificity largely depends on the combination of proteins incorporated into their complexes56. HDAC1 and 2 associate with Sin3 scaffolded complexes which serve a range of functions within the cell. The substrate specificity and function of these HDAC isozymes can change by altering the protein composition of the complex57. Although HDAC8 is phylogenetically most similar to the other class I HDACs, divergent evolution12 may have altered how HDAC8 interacts with cofactors, possibly allowing this isozyme to function independent of other proteins. However, HDAC8 does associate with other proteins, and these interactions likely affect the biological function and selectivity of this enzyme.
Distinguishing between HDAC8 substrates and binding partners in the cell is currently difficult, as discussed in the previous sections. For example, previous experiments have provided evidence that the HDAC1/HDAC2 complex associates with both the PP1 phosphatase and CREB, leading to decreased CREB phosphorylation52. Because an inactive HDAC1 mutant still affects CREB activity, the function of the HDAC1/HDAC2 complex was proposed to co-localize PP1 phosphatase and CREB. However, it is possible that HDAC2 catalyzes deacetylation of CREB under these conditions52. Similarly, both PP1 and CREB co-immunoprecipitate with HDAC8, and HDAC8 overexpression decreases CREB activity. These data are consistent with HDAC8 either acting as a scaffold to enhance the interaction between PP1 phosphatase and CREB or catalyzing deacetylation of CREB.
HDAC8 also co-localizes with α-actin, as indicated by immunofluorescence staining28. This interaction was confirmed by pulldown experiments using human smooth muscle cells, demonstrating an endogenous association between α-actin and HDAC829,43. The function of this interaction was partially elucidated by demonstrating that siRNA knockdown of HDAC8 in human smooth muscle cells decreased the ability of cells to contract, when exposed to a collagen lattice. Furthermore, the siRNA-treated smooth muscle culture cells were smaller and unable to spread. These changes in cell morphology occurred without detectable changes to α-actin acetylation29, suggesting that HDAC8 acts as part of a complex which modulates the cell cytoskeleton. Furthermore, pulldown experiments demonstrate that HDAC8 associates with the proteins Hsp20, myosin heavy chain, and cofilin43 all of which can potentially affect actin dynamics58–59. It is currently unclear whether Hsp20 or cofilin are acetylated and substrates for HDAC8. However, HDAC8 associates better with the non-acetylated form of myosin heavy chain, suggesting that this protein is not an HDAC8 substrate43. Because HDAC8 enhances cell contractility and associates with three proteins important for actin function, it is likely that HDAC8 is a component of a complex that modulates actin dynamics.
Additional potential HDAC8 interaction partners have been identified using a bacterial two-hybrid system60. Two of the fifteen identified binding partners have been examined in detail: the human Ever-Shorter Telomeres 1B (hEST1B) protein that activates telomerase activity and HOP1, an adaptor protein linking Hsp70 and Hsp90. The two-hybrid results were confirmed using co-immunoprecipitation of overexpressed hEST1B and HDAC8 in Hela cells. HDAC8 knockdowns led to decreased telomerase activity through diminished levels of hEST1B. As HDAC8 activity does not affect the promoter region regulating hEST1B, the hEST1B level is likely not regulated by alteration in transcription. However, hEST1B levels are increased by addition of a proteasome-dependent pathway inhibitor or decreased by overexpression of ubiquitin and rescued by phosphorylated HDAC8. These results argue that phosphorylated HDAC8 protects hEST1B from polyubiquitination and subsequent degradation by the proteosome. The protective effects of phosphorylated HDAC8 on hEST1B levels are independent of deacetylase activity, remaining in the presence of the catalytically inactive His143Ala-HDAC8 mutant, or after exposure of cells to TSA. Therefore, HDAC8 interacts with hEST1B but deacetylation is not required for the functional effect. To further explore the interaction between HDAC8 and HOP1 indicated by the two-hybrid experiment, the association of HDAC8 with known HOP1 binding partners was investigated. Pulldown experiments demonstrated that endogenous Hsp70 and Hsp90 co-immunoprecipitate with overexpressed HDAC860. This result suggests that HDAC8, HOP1, Hsp70, and Hsp90 form a complex. One proposed mechanism for the effect of HDAC8 on telomerase activity suggests that the Hsp70-HDAC8 complex protects hEST1B from ubiquitination catalyzed by the E3 ubiquitin ligase CHIP60. This in turn raises the levels of hEST1B and activates telomerase. Interestingly, interaction of HDAC8 with the Hsp proteins may help to elucidate the effect of HDAC8 on α-actin, since Hsp90 has been proposed to modulate α-actin dynamics61–62. Thus it is possible that the HDAC8-HOP1-Hsp90 complex may regulate α-actin function.
Enzyme structure affects substrate specificity
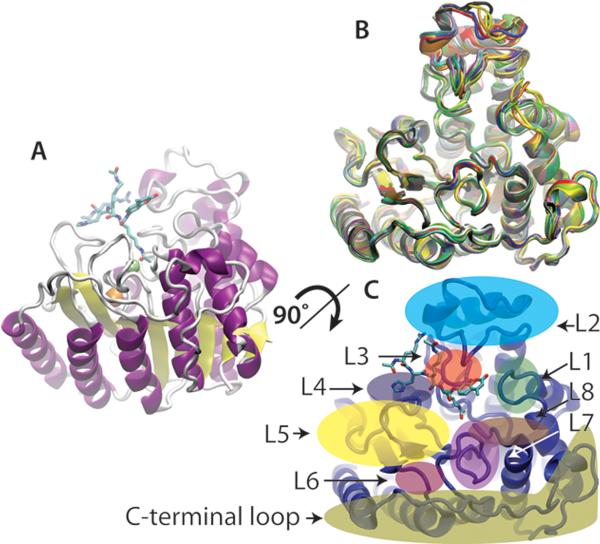
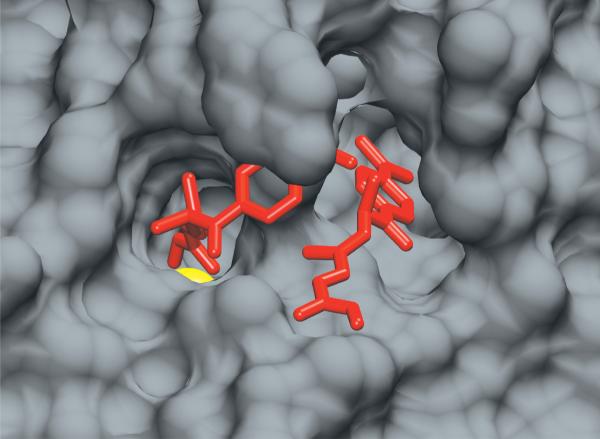
The structure of HDAC8 yields clues about molecular recognition relevant to substrate selectivity. HDAC8 is the second smallest metal-dependent HDAC at ~42 kDa, containing little more than the catalytic domain12,19–21. This HDAC folds as a single α/β domain with a core eight-stranded β-sheet surrounded by eleven α-helices [figure 3a]. The substrate binding surface, composed of nine loops and an 11Å tunnel leading to the active site, is proposed to be conformationally flexible based on the poor occupancy and varying positions of the loop residues in crystal structures54–55,63–67 [Figure 3b]. Furthermore, one crystal structure illuminates a bound TSA molecule interacting with residues in the hydrophobic core of HDAC8 64 [Figure 4]. While this may simply be an artifact, the alternative binding mode suggests that the surface of the protein can change conformation enough to allow hydrophobic molecules to intercalate between these loops and interact with the interior of the protein. Loops are a common structure in promiscuous enzymes68 and examples of proteins, such as chymotrypsin69 and carboxypeptidase A70, that use loops to bind a range of substrates are abundant in nature. These loops create a number of different conformations that bind ligands through a combination of induced fit and select fit mechanisms31,71. The varied conformations and motifs provide a palette of binding sites to accommodate a multiplicity of substrates. Furthermore, long range allosteric movements propagated through the loops may affect the active site and surrounding areas, potentially altering substrate preferences.
Figure 3.
HDAC8 structures. A. PDBID: 2v5w63. Side view of HDAC8 with bound peptide substrate. Helices are purple, sheets are yellow, turns are white, the monovalent cations are orange, and the active site metal is colored green. The Fluor de lys substrate representing the p53 sequence is colored cyan for carbon, red for oxygen, and blue for nitrogen. B. Front view of an overlay of the 21 HDAC8 crystal structures in the PDB: PDBID: 2v5x, 2v5w, 1t69, 1t64, 1vkg, 1t67, 1w22, 3sfh, 3sff, 3mz3, 3ezt, 3fo6, 3mz4, 3mz6, 3mz7, 3ew8, 3ezp, 3f07, 3f0r, 3ewf, and 3rqd54–55,63–67. Structural variations are especially apparent in the L1, L2, and C-terminal loops. C. A map of the crystal structure of HDAC8 outlining the loop regions.
Figure 4.
HDAC8 with two bound TSA molecules. PDBID: 1t6464. In this crystal structure, one molecule of TSA binds to the active site tunnel to coordinate the divalent metal ion (colored yellow) while a second TSA molecule binds nearby in between the L1, L2, and L3 loops.
In fourteen of the twenty-one HDAC8 crystal structures, the enzyme crystallizes as a dimer along the substrate binding interface54–55,63–66. As HDAC8 is a monomer in solution67, the dimer interface may provide insight into long range interactions between HDAC8 and its in vivo substrates. To date, substrate specificity has mainly been evaluated using peptide substrates, therefore only short range interactions have emerged as HDAC8 substrate binding motifs22–24,33. Based on the crystal structure of bound peptides55,63 and biochemical measurements, these interactions include ring stacking, hydrogen bonding, salt bridges, and electrostatic interactions. Ring stacking between Tyr100 and the methylcoumarin of the Fluor-de-lys peptides is observed in the crystal structure55,63. Similarly, ring stacking between aromatic amino acids in the +1 position and Tyr100 may be important for substrate recognition23,33. Additionally, hydrogen bonding between the backbone amides of the substrate and the Asp101 side chain oxygens are important for molecular recognition55. Salt bridges between positively charged arginines in the substrate and negatively charged carboxylate side chain oxygens, and general hydrophobic interactions can be seen in the peptide-enzyme interface55,63. Because of the limited number of interactions, the binding affinity may be dominated by a few strong contacts, as observed for the interaction between Tyr100 of HDAC8 and the methylcoumarin moiety of short Fluor-de-lys peptides33. This pi-pi interaction (~2 kcal/mol)72–73 is of comparable energy with other HDAC8-peptide contacts. In contrast, binding a protein substrate could involve many more contacts, including multiple hydrogen bonds (0.5–1.5 kcal/mol), hydrophobic (~1 kcal/mol), electrostatic (<1 kcal/mol)31, and solvent exposed salt bridge (~1–3 kcal/mol)74 interactions. Therefore, the binding affinity could depend on a large number of interactions that together create a promiscuous substrate binding profile. Determinants of substrate specificity are still being evaluated for HDACs and further identification of binding motifs will be beneficial for understanding the biology of these enzymes.
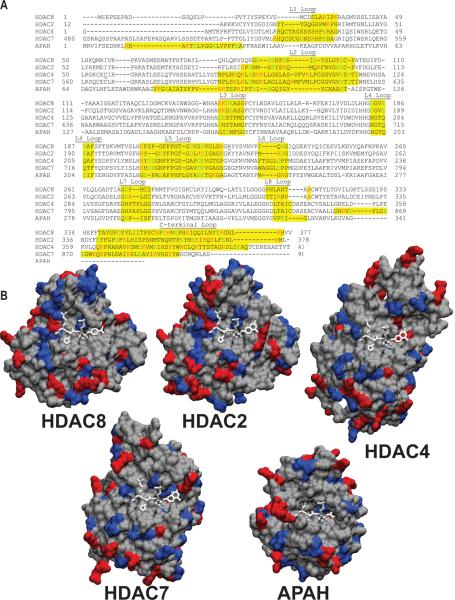
When the structure of HDAC8 is compared to that of the homologous polyamine deacetylase, APAH75, striking differences in loop size and structure can be observed. These differences in the loops may be important for substrate binding as APAH catalyzes deacetylation of small molecules, including acetylated spermidine, putrescine, and spermine, while HDAC8 deacetylates macromolecules. In APAH, the L1 and L2 loops are much larger and contain many more hydrophobic residues than in the corresponding HDAC8 loops [Figure 5a,b], while the C-terminal loop and helix in HDAC8 are absent in APAH. Similarly, a comparison of the L1, L2, and C-terminal loops of different HDACs reveals interesting variations. The L1 and L2 loops of HDAC276, 477, 778, and 863 are more divergent in size, structure, and number of charged residues than other loops within these HDACs [Figure 5a, b]. For instance, the size and number of charges within the L1 and L2 loops change two-fold between HDAC8 and HDAC4. Comparison of all the HDAC8 crystal structures illustrates that the L1 and L2 loops have the most structural variability of the loops in the proposed substrate binding surface, suggestive of a role in ligand binding. Additionally, the L2 loop interacts with inhibitors, suggesting that it may be important for molecular recognition of substrates55. The L3 loop, which lies below the L2 loop and flanks the active site, also varies greatly in the number of charges in the loop among HDACs 2, 4, 7, and 8, consistent with a role in substrate or binding partner selectivity. The C-and N- terminal portions of the HDACs, which lie on the outer edge of the substrate binding surface, may also interact with ligands. In the HDAC crystal structures, the C-terminal loops vary in position, charge, and size and may be responsible for long distance interactions between HDACs and their substrates, or used for recognition of binding partners.
Figure 5.
Structural variation within the structurally characterized HDACs. A. Aligned sequences of the published HDAC crystal structures63,75–78. Highlighted in yellow are the residues that comprise the loop regions and amino acids that comprise the putative substrate binding region. The positively charged residues are red and the negatively charged residues are green. B. Surface visualizations of the crystal structures for HDAC2 (PDBID: 3max76), HDAC4 (PDBID: 2vqm77), HDAC7 (PDBID: 3c0y78), HDAC8 (PDBID: 2v5w63), and APAH (PDBID: 3q9b75). Superimposed into each structure is the Fluor de lys substrate (white) from the HDAC8 structure. In red are the positively charged residues Arg and Lys, and in blue are the negatively charged residues Asp and Glu.
Along with structural studies, peptide substrates have been useful for evaluating substrate motifs recognized by HDAC8. Reister and colleagues measured the reactivity of HDAC8 with a peptide library of the sequence Ac-X-Z-K(ac)-methylcoumarin, where X and Z were all amino acids except for cystine22. This work indicated that HDAC8 favors Pro, Met, Ala, Lys, Arg, Gln, Asp, Phe, and Ser at the −2 position and aromatic (Phe, Trp, and Tyr) and hydrophobic (Ile, Met, and Val) amino acids at the −1 position. However, the activity of HDAC8 in these assays was low, possibly due to the inclusion of the metal chelator EDTA in the assay. The Mrksich group developed a mass spectrometric assay to profile the local substrate specificities of HDACs 33. The reactivity of HDAC8 with a peptide array of the sequence, Ac-G-X-K(ac)-Z-G-C-NH2 where X and Z were any amino acid other than cysteine, showed that the most efficient substrate contains Arg and Phe at the X and Z positions, respectively33. However, HDAC8 also catalyzes deacetylation of peptides containing the sequence X=Arg/Z= variable and X = variable/Z = Phe. HDAC8 selectivity was further screened using a peptide library with the following sequence: Ac-G-R-K(ac)-X-Z-C-NH223. These data demonstrated a preference for Arg or Phe at the X position. Furthermore, when X is Phe the identity of the Z position has only a modest effect on activity. These results suggest that specific positions and combinations of amino acids contribute significantly to the substrate recognition of small peptides, while other positions fine tune recognition. The Mrksich group also demonstrated that an RHR motif added to the C-terminus of peptide substrates of varying lengths enhances reactivity, demonstrating that distal sequences can modulate HDAC8 substrate selectivity24. Interestingly, the sequences RHRK and RHKK are found in the H4 histone tail and in p53, respectively, and hint that distal sequences may enhance the reactivity of HDAC8 with these substrates in cells.
Finally, the structure of the active site may also play a role in HDAC substrate specificity. HDAC2 and 8 have well defined 11Å channels leading to their active sites that easily accommodate an acetyllysine side chain, however, this tunnel is lacking in HDAC4 and 7 63,76–78 where only half of the channel is apparent. This modification in active site structure could suggest that HDAC4 and 7 catalyze deacetylation of alternate substrates, as proposed by Lombardi et. al.75. Alternatively, these isozymes might need substrates that complement the active site to stabilize the binding of the acetyllysine moiety.
Catalytic Mechanism and Regulation of HDAC8 activity
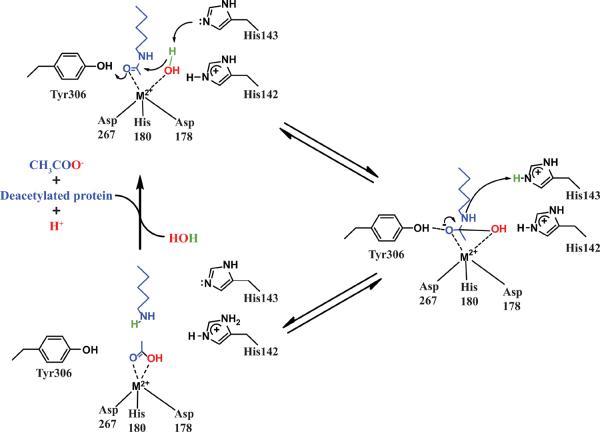
The active site of HDAC8 contains a divalent metal ion coordinated to two aspartate and one histidine side chain (Asp178, Asp267, and His180) and one or two water molecules. Additionally, a conserved tyrosine (Tyr306) and a pair of conserved histidine/aspartate hydrogen bond dyads (His142/Asp176 and His143/Asp183) are located near the bound acetyllysine moiety (Fig. 6). The enzyme is proposed to catalyze hydrolysis using a metal-coordinated water nucleophile and general acid-base catalysis (GABC) with either one or two side chains, similar to typical metallohydrolase mechanisms [Figure 6]13,55,79–80. The substrate binds to HDAC8 with the catalytic metal coordinating both the carbonyl oxygen of the acetyllysine substrate and a water molecule. In the first step of the mechanism, His142 functions as a general base to abstract a proton from the metal-bound water, as this nucleophile reacts with the carbonyl carbon to form a high energy tetrahedral intermediate. The oxyanion intermediate is proposed to be stabilized by coordination with the metal ion, hydrogen bonding with Tyr306, and electrostatic interactions with positively charged groups in the active site. Proton donation from an active site general acid to the amine leaving group accompanies breakdown of the tetrahedral intermediate to form the acetate and the deacetylated lysine products79. In the GABC mechanism originally proposed from the crystal structure of the homologous HDLP enzyme79, His142 and protonated His143 are proposed to function as the general base and general acid, respectively. In the one GABC mechanism, H143 functions as both the general acid and general base catalyst and H142 acts as an electrostatic catalyst55,80, similar to the mechanism proposed for carboxypeptidase A13. Subsequent studies utilizing mutagenesis and molecular dynamics simulations suggest a preference for the one base mechanism55,80,30.
Figure 6.
Schematic of the one base mechanism for HDAC8. Blue is the acetyl-lysine of the substrate while the nucleophilic water is green and red. For clarity, equilibration of exchangeable protons with solvent is not shown.
The HDAC8 crystal structure also contains two monovalent cation sites54–55,63–66, suggesting that the activity of HDAC8 may be modulated by both the concentration and type of ions in solution. One monovalent cation site is 7 Å from the divalent catalytic metal ion and is coordinated by the side chain oxygens of Asp176 and Ser199 and the backbone carbonyl oxygens of Asp176, Asp178, His180, and Leu200. The second site is 21 Å from the divalent catalytic metal ion, and is ligated by two water molecules and the backbone carbonyl oxygens of Phe189, Thr192, Val195, and Tyr225. Initial activity measurements demonstrated that the concentrations of K+ and Na+ modulate HDAC8 catalysis [biolmol unpublished]. A detailed examination demonstrated that the value of kcat/KM for HDAC8-catalyzed deacetylation has a biphasic dependence on the concentration of K+ and Na+ ions40. In the absence of monovalent ions, the activity of HDAC8 is very low; addition of monovalent cations to Zn-bound HDAC8 increases activity with K1/2,act = 14 mM for K+. At higher K+ concentrations Zn-HDAC8 activity is inhibited with K1/2,inhib = 130 mM. Mutagenesis studies indicate a significant decrease in potassium inhibition in the His142Aala and Asp176Ala/Asn mutants indicating that the monovalent ion site near the active site is inhibitory. Potassium binding next to His142 has been proposed to lower the pKa of this residue, decreasing the concentration of protonated His142, thereby lowering catalytic activity. Similar biphasic regulation has been measured for Na+, but activation and inhibition require a five-fold and ten-fold higher concentration of Na+ compared to K+, respectively40. At the 100 mM K+ concentration within smooth muscle cells81, HDAC8 activity is partially inhibited and sensitive to changes in the K+ concentration.
HDAC8 catalytic activity is enhanced by a number of divalent metal ions, including Co2+, Zn2+, Ni2+, and Fe2+ 18. When HDAC8 is purified under aerobic conditions, the bound metal ion is Zn2+. However, recombinant HDAC8 purified anaerobically from E. coli contains 8-fold more iron than zinc, consistent with this, the recombinant HDAC8 activity in E. coli cell lysates is oxygen-sensitive18. Additionally, although HDAC8 binds Zn2+ nearly 106-fold more tightly than Fe2+ 54, the affinities for both metal ions are comparable to the readily exchangeable metal concentrations estimated in living cells, suggesting that HDAC8 can bind either Fe2+ or Zn2+ in vivo. Furthermore, the identity of the bound metal ion alters the catalytic properties of HDAC8. When catalyzing deacetylation of the methylcoumarin-labeled p53 peptide, the kcat/KM value for Fe2+-bound HDAC8 is almost three times larger than that of Zn2+-HDAC8. Interestingly, substitution of Fe2+ for Zn2+ also decreases the values of KM and KI for SAHA, suggesting that Fe2+ enhances ligand affinity18. However, a comparison of the crystal structures of the hydroxamate-bound Fe2+-HDAC8 and Zn2+-HDAC8 shows no significant differences in the active site or the rest of the protein54. These data suggest that either binding of the hydroxamic inhibitor stabilizes a common enzyme conformation, or that the bound metal ion affects protein dynamics that are not observable by crystallography.
Comparison of the Zn2+/Fe2+ metal affinities with the cellular concentrations of those metals suggests that HDAC8 likely binds a combination of iron and zinc cofactors in eukaryotic cells54. Furthermore, the cellular zinc concentration can change dramatically upon oxidative stress82–83 and metal toxicity84 potentially altering the populations of Fe2+-HDAC8 and the Zn2+-HDAC8 based on cellular conditions. This provides a means by which the cell can couple HDAC8 activity to cellular stresses.
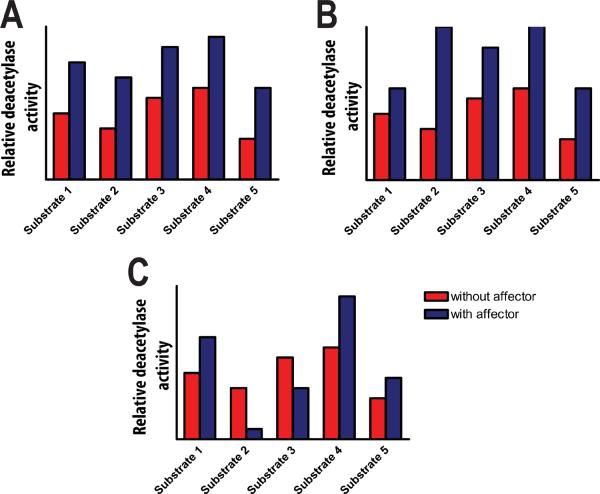
A simple model for HDAC activation and inhibition assumes that compounds, cofactors, and binding partners equally affect the activity of HDAC8 with all substrates [Figure 7a]. An alternative to this model proposes that substrate selectivity may be differentially regulated by stimuli. For example, scaffolding activators could preferentially enhance the binding of HDAC8 to one set of substrates [Figure 7b]. Similarly, alteration of the active site metal ion or bound monovalent ions could alter ligand specificity. For example, Fe2+-HDAC8 binds the inhibitor SAHA 2-fold more tightly than Zn2+-HDAC840 even though Zn2+ is a stronger Lewis acid85.
Figure 7.
Schematic of three potential models for describing the effect of an activating effector on HDAC activity. A. In this model catalysis of deacetylation of each substrate is enhanced by an equivalent factor upon addition of the effector. B. In this model catalysis of deacetylation of each substrate is enhanced by a different factor upon addition of the effector. C. In this model catalysis of deacetylation of some substrates is activated while other substrates are inhibited by the effector.
This change in binding affinity suggests that the active site metal ion may contribute subtly to the structure, dynamics, and molecular recognition of HDACs.
HDAC8 localization
Most simply, protein localization may regulate HDAC8 substrate specificity by changing the effective substrate concentration. HDACs have been found to have a range of cellular locations. HDAC1 and 2 are exclusively nuclear, while HDAC6 is mostly cytoplasmic, and HDAC3, 4, 5, 6, 7, 9, 10, and 11 appear to shuttle in and out of the nucleus86. Initially, HDAC8 was found to have a putative nuclear localization site and was observed in the nucleus of NIH3T319 and HEK293 cells20. Soon after, microscopy demonstrated that HDAC8 localizes to both the cytoplasm and nucleus of embryonic smooth muscle cells, skin fibroblasts, and NIH3T3 cells28 although there remains some skepticism about this point. HDAC3, the closest HDAC8 human homologue12, exists in both the cytoplasm and nucleus, and localization has been linked to the regulation and cellular function of this enzyme. Whether cellular localization plays a role in HDAC8 activity is currently unknown, as no studies have yet broached this subject.
Determining the cell type-dependent expression of HDAC8 may provide interesting insights about its substrate specificity and biological function. In general, class I HDACs are ubiquitously expressed amongst the various cells of an organism, while class II HDACs are more cell-type specific86–87. Likewise, HDAC8 has been found in a number of different healthy and diseased cell types [see supplemental table].
HDAC8 knockouts after birth are non-lethal88, consistent with the ability of humans to tolerate pan-HDAC inhibitors as an anti-cancer treatment89. However, protein expression profiles can vary significantly during development and several HDAC knockouts are lethal during mammalian embryonic development8. For example, cells lacking HDAC3 die before embryonic day 9.5; deletion of HDAC3 leads to hyperactivity of the nuclear receptor PPARα and problems with embryonic gastrulation90. Similarly, HDAC8 expression is crucial to development, as mice lacking this enzyme die soon after birth88. Death is due to brain hemorrhaging caused by defects in the development of the mouse skull resulting from problems with neural crest patterning. These skull defects are similar to those that occur upon overexpression of the transcription factors Otx2 and Lhx1, suggesting that HDAC8 either directly regulates these proteins or affects regulators of these proteins88. The mechanism of HDAC8 regulation of Otx2 and Lhx1 has yet to be determined. Furthermore, since HDAC8 knockouts are not lethal after birth88, it is unclear whether HDAC8 no longer regulates these proteins, whether this regulation still occurs but is not vital for viability, or another mechanism takes place.
Post-translational modification of HDAC8
Post-translational modifications such as phosphorylation, may also regulate HDAC8 activity. A screen of three protein kinases, casein kinase II, protein kinase A (PKA) and protein kinase G (PKG) indicated that HDAC8 phosphorylation could be catalyzed by both PKA and PKG91. PKA phosphorylation appeared to be predominant and this function was authenticated in vivo by incubation of cells with the PKA inhibitor H-89, which lowered HDAC8 phosphorylation levels92. Based on consensus sequences, nineteen potential phosphorylation sites were identified in HDAC8. Phosphoamino acid analysis followed by two dimensional thin layer chromatography demonstrated modification of a serine residue92 and, based on this information, Ser39 was identified as the only PKA phosphorylation site in the HDAC8 sequence19,92. A Ser39Ala HDAC8 mutant, which cannot be phosphorylated, negates phosphorylation of HDAC8 catalyzed by PKA, confirming this location as the primary phosphorylation site on HDAC8. Furthermore, phosphorylation of this site modulates HDAC8 activity. The specific activity of HDAC8 purified from cells treated with forskolin, a PKA activator, decreased by five-fold in an in vitro assay using purified histones92. Furthermore, the specific activity of Ser39Glu HDAC8, a mutation that mimics phosphorylation, decreases to a level comparable to that of phosphorylated HDAC8, while the specific activity of the Ser39Ala mutant is similar to unmodified HDAC8. To examine whether in vivo effects of phosphorylation of HDAC8 correlate with the in vitro measurements, HDAC8-transfected HeLa cells were treated with forskolin. These cells showed increased levels of acetylated histones H3 and H4, suggesting that the decreased deacetylase activity of phosphorylated HDAC8 led to increased acetylation in vivo92.
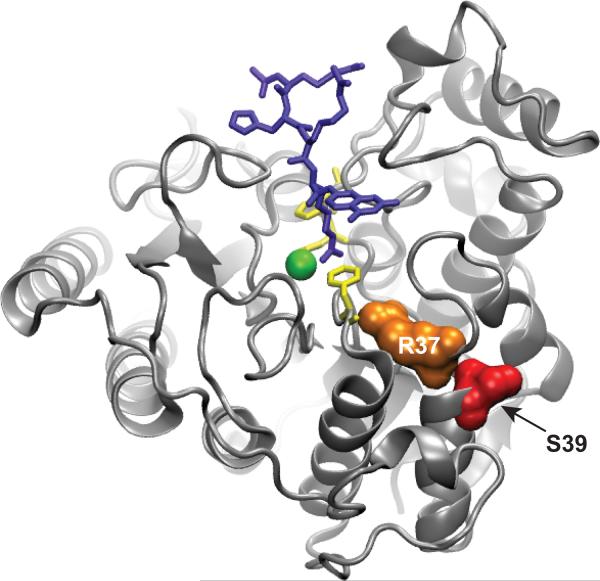
Ser39 is located on the backside of the HDAC8 surface, 21Å from the catalytic metal ion54–55,63–66 [Figure 8]. Nonetheless, phosphorylation has the potential to affect the subcellular localization, protein-protein interactions, allosteric effects, and HDAC8 activity via conformational changes that propagate to the active site or enzyme-substrate interface. Ser39 lies near the junction with the L1 loop54–55,63–66 that has been implicated in substrate recognition, and therefore phosphorylation at that position may alter enzyme-substrate interactions. The Ser39 residue is located in a pocket on the enzyme surface surrounded by hydrophobic and acidic residues suggesting that phosphorylation of Ser39 could induce a drastic structural perturbation due to charge repulsion64. Ser39 also directly contacts the conserved Arg37 residue, which is proposed to be important for gating an acetate release channel in HDAC853 [Figure 8]. The Arg37Ala mutation decreases the kcat/KM value for Co2+-HDAC8-catalyzed deacetylation of the Fluor-delys substrate (R-H-K(Ac)-K(Ac)-fluorophore) by 530-fold53. Based on the proximity of Ser39 to Arg37, phosphorylation at this position may similarly affect HDAC8 activity.
Figure 8.
Phosphorylation of Ser39 may affect the active site structure and/or reactivity of HDAC8. PDBID: 2v5w 63. This structure shows that phosphorylation of Ser39 (red) may be able to perturb the position and/or electrostatic environment of Arg37 (orange) and in turn, affect the active site residues (yellow). Blue is the Fluor de lys substrate and green is the active site metal.
Phosphorylation may also regulate HDAC8 through the modulation of protein-protein interactions. In the bacterial two-hybrid assay that identified fifteen HDAC8-interacting proteins60 expression of PKA was necessary for the pulldown of six of these identified proteins, and suggests that these proteins interact solely with phosphoHDAC8. Two of these interactions, those between HDAC8 and hEST1B and between HDAC8 and Hsp70, were further observed by co-immunoprecipitation, showing that treatment of cells with forskolin, led to increased amounts of phosphorylated HDAC8 and increased interactions60. These data strongly suggest that HDAC8 phosphorylation regulates HDAC8 complex formation. Similarly, phosphorylation of HDAC1 and HDAC2 regulates association of these proteins with complexes such as mSin3A, RbAp48, and CoREST91,93. Phosphorylation-dependent complex formation may also regulate the cellular localization of HDAC8. Fluorescence microscopy of myometrial cells shows that HDAC8 and phosphoHDAC8 both localize primarily to the cytosol, but cell fractionation data suggest that phosphoHDAC8 has increased association with the cytoskeleton compared to HDAC8 in this cell type43. HDAC4, HDAC5, and HDAC7 have been proposed to utilize nuclear-cytoplasmic shuttling mechanisms involving phosphorylation-dependent binding to 14-3-3 proteins for regulating their subcellular localization, and a similar mechanism may regulate HDAC8 localization94–97.
The Ser39 site is an interesting location for phosphorylation amongst HDACs. Ser39 is not conserved among class I HDACs; the residue in the corresponding position of other class I HDACs is arginine in HDAC1 and 2, and alanine in HDAC3. Also, HDAC8 and HDAC5 contain the only phosphorylation sites that are located within the HDAC catalytic domain98,99–100. Additionally, HDAC8 is the only isozyme phosphorylated by PKA100. In general, the effect of phosphorylation on the activity of other class I isozymes, HDAC1 and 2, is ambiguous and/or contradictory91,93,101–102. For example, phosphorylation of HDAC1 had little to no effect on deacetylase activity using a synthetic histone H4 peptide101–102 but activity on isolated histones decreased using mutants that could not be phosphorylated93. Therefore HDAC8 may be the best isozyme for examining the role of phosphorylation in regulating acetylation.
Many HDACs undergo additional post-translational modification including acetylation, ubiquitination, and sumoylation99, but additional modifications of HDAC8 have not yet been demonstrated. HDAC8 has a consensus motif for glycosylation at Asn136 that could be modified19,92; however the NetNGlyc 1.0 server does not predict N-glycosylation of this site due to the lack of a signal peptide103. Acetylation has been observed for HDAC1 at multiple sites, and one of the acetylated residues is conserved in HDAC8. Two of the HDAC1 sites are located in the deacetylase domain and four sites are near the C-terminus; acetylation of these sites inhibits HDAC1 deacetylase activity toward histones in vitro and corepressor function in vivo104. The two sites in the deacetylase domain, Lys218 and Lys220, are located near the activating monovalent cation binding site, so decreased activity from acetylation of these residues may arise from alteration of monovalent cation binding104. Sequence alignment by Cobalt indicates that the Lys218 position in HDAC1 is conserved in the corresponding Lys221 position in HDAC8 [(http://www.ncbi.nlm.nih.gov/tools/cobalt/)]. As this monovalent site activates HDAC8 allosterically40, it is feasible that HDAC8 activity could be regulated by modification at this location. However, no modifications at this site have yet been observed and post-translational modifications of HDAC8 need to be further examined.
Concluding remarks
Due to the abundance and vital function of acetylation within the cell, enzymes that catalyze acetylation and deacetylation are regulated in a multitude of ways and on a number of time scales. One mechanism of regulating HDAC activity is changing the substrate preferences for these enzymes, which in turn affects cellular processes. These regulatory mechanisms may allow the cell to finely tune the substrate preference for many HDACs simultaneously by allowing the same stimuli to differentially alter the activity of each HDAC isozyme. Understanding the interplay between various stimuli and HDAC regulation will give us tremendous insight into the inner workings of cellular processes and the mechanisms of disease formation. Even though HDAC8 has been extensively studied, it is humbling to know the vast amounts of information that have yet to be determined regarding the cohort of HDAC8 substrates and binding partners, localization in the cell, and regulatory mechanisms. Therefore, even for the best-characterized HDAC, there are likely many factors that affect substrate recognition that have not yet been discovered. The dissection of these factors in the future will be tremendously important for understanding not only the cellular function of HDACs, but also cellular regulation by post-translational modifications.
Supplementary Material
Acknowledgements
We thank Dr. Patrick O'Brien for his useful insight into the paper's contents, Tony Mustoe for his help with visualization of molecules, Mark Taylor, Felicia Gray, Alison Tebo, and members of the Fierke Lab for their critical reading of this paper and discussion. This material was supported, in whole or in part, by the Nation Institutes of Health Grants, NIGMS GM40602 (CAF), T32-GM-008353, and 5-T32-GM-008597. This material is based upon work supported by the National Science Foundation under Grant No. DGE0718128.
References
- 1.Yang XJ, Seto E. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glozak MA, Sengupta N, Zhang X, Seto E. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Workman JL, Kingston RE. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 4.Glozak MA, Seto E. Oncogene. 2007;26:5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 5.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 6.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khoury GA, Baliban RC, Floudas CA. Sci Rep. 2011;1 doi: 10.1038/srep00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haberland M, Montgomery RL, Olson EN. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kazantsev AG, Thompson LM. Nat Rev Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 10.Drummond DC, Noble CO, Kirpotin DB, Guo Z, Scott GK, Benz CC. Annu Rev Pharmacol Toxicol. 2005;45:495–528. doi: 10.1146/annurev.pharmtox.45.120403.095825. [DOI] [PubMed] [Google Scholar]
- 11.Spiegel S, Milstien S, Grant S. Oncogene. 2012;31:537–551. doi: 10.1038/onc.2011.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregoretti IV, Lee YM, Goodson HV. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Hernick M, Fierke CA. Arch Biochem Biophys. 2005;433:71–84. doi: 10.1016/j.abb.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Frye RA. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 15.Milne JC, Denu JM. Curr Opin Chem Biol. 2008;12:11–17. doi: 10.1016/j.cbpa.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 16.Durst KL, Lutterbach B, Kummalue T, Friedman AD, Hiebert SW. Mol Cell Biol. 2003;23:607–619. doi: 10.1128/MCB.23.2.607-619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson BJ, Tremblay AM, Deblois G, Sylvain-Drolet G, Giguere V. Mol Endocrinol. 2010;24:1349–1358. doi: 10.1210/me.2009-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gantt SL, Gattis SG, Fierke CA. Biochemistry. 2006;45:6170–6178. doi: 10.1021/bi060212u. [DOI] [PubMed] [Google Scholar]
- 19.Hu E, Chen Z, Fredrickson T, Zhu Y, Kirkpatrick R, Zhang GF, Johanson K, Sung CM, Liu R, Winkler J. J Biol Chem. 2000;275:15254–15264. doi: 10.1074/jbc.M908988199. [DOI] [PubMed] [Google Scholar]
- 20.Buggy JJ, Sideris ML, Mak P, Lorimer DD, McIntosh B, Clark JM. Biochem J. 2000;350(Pt 1):199–205. [PMC free article] [PubMed] [Google Scholar]
- 21.Van den Wyngaert I, de Vries W, Kremer A, Neefs J, Verhasselt P, Luyten WH, Kass SU. FEBS Lett. 2000;478:77–83. doi: 10.1016/s0014-5793(00)01813-5. [DOI] [PubMed] [Google Scholar]
- 22.Riester D, Hildmann C, Grunewald S, Beckers T, Schwienhorst A. Biochem Biophys Res Commun. 2007;357:439–445. doi: 10.1016/j.bbrc.2007.03.158. [DOI] [PubMed] [Google Scholar]
- 23.Gurard-Levin ZA, Kilian KA, Kim J, Bahr K, Mrksich M. ACS Chem Biol. 2010;5:863–873. doi: 10.1021/cb100088g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gurard-Levin ZA, Mrksich M. Biochemistry. 2008;47:6242–6250. doi: 10.1021/bi800053v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dose A, Liokatis S, Theillet FX, Selenko P, Schwarzer D. ACS Chem Biol. 2011;6:419–424. doi: 10.1021/cb1003866. [DOI] [PubMed] [Google Scholar]
- 26.Juan LJ, Shia WJ, Chen MH, Yang WM, Seto E, Lin YS, Wu CW. J Biol Chem. 2000;275:20436–20443. doi: 10.1074/jbc.M000202200. [DOI] [PubMed] [Google Scholar]
- 27.Luo J, Su F, Chen D, Shiloh A, Gu W. Nature. 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 28.Waltregny D, De Leval L, Glenisson W, Ly Tran S, North BJ, Bellahcene A, Weidle U, Verdin E, Castronovo V. Am J Pathol. 2004;165:553–564. doi: 10.1016/S0002-9440(10)63320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waltregny D, Glenisson W, Tran SL, North BJ, Verdin E, Colige A, Castronovo V. FASEB J. 2005;19:966–968. doi: 10.1096/fj.04-2303fje. [DOI] [PubMed] [Google Scholar]
- 30.Gantt SL. Chemistry. The University of Michigan; Ann Arbor: 2006. [Google Scholar]
- 31.Fersht A. Structure and mechanism in protein science : a guide to enzyme catalysis and protein folding. W.H. Freeman; New York: 1999. [Google Scholar]
- 32.Wegener D, Wirsching F, Riester D, Schwienhorst A. Chem Biol. 2003;10:61–68. doi: 10.1016/s1074-5521(02)00305-8. [DOI] [PubMed] [Google Scholar]
- 33.Gurard-Levin ZA, Kim J, Mrksich M. Chembiochem. 2009;10:2159–2161. doi: 10.1002/cbic.200900417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schultz BE, Misialek S, Wu J, Tang J, Conn MT, Tahilramani R, Wong L. Biochemistry. 2004;43:11083–11091. doi: 10.1021/bi0494471. [DOI] [PubMed] [Google Scholar]
- 35.Riester D, Wegener D, Hildmann C, Schwienhorst A. Biochem Biophys Res Commun. 2004;324:1116–1123. doi: 10.1016/j.bbrc.2004.09.155. [DOI] [PubMed] [Google Scholar]
- 36.Smith BC, Denu JM. J Biol Chem. 2007;282:37256–37265. doi: 10.1074/jbc.M707878200. [DOI] [PubMed] [Google Scholar]
- 37.Bar-Even A, Noor E, Savir Y, Liebermeister W, Davidi D, Tawfik DS, Milo R. Biochemistry. 2011;50:4402–4410. doi: 10.1021/bi2002289. [DOI] [PubMed] [Google Scholar]
- 38.Alarcon R, Orellana MS, Neira B, Uribe E, Garcia JR, Carvajal N. FEBS J. 2006;273:5625–5631. doi: 10.1111/j.1742-4658.2006.05551.x. [DOI] [PubMed] [Google Scholar]
- 39.Villena JA, Kralli A. Trends Endocrinol Metab. 2008;19:269–276. doi: 10.1016/j.tem.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gantt SL, Joseph CG, Fierke CA. J Biol Chem. 2010;285:6036–6043. doi: 10.1074/jbc.M109.033399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitelman F, Heim S. Genes Chromosomes Cancer. 1992;5:57–66. doi: 10.1002/gcc.2870050109. [DOI] [PubMed] [Google Scholar]
- 42.Liu P, Tarle SA, Hajra A, Claxton DF, Marlton P, Freedman M, Siciliano MJ, Collins FS. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 43.Karolczak-Bayatti M, Sweeney M, Cheng J, Edey L, Robson SC, Ulrich SM, Treumann A, Taggart MJ, Europe-Finner GN. J Biol Chem. 2011;286:34346–34355. doi: 10.1074/jbc.M111.278549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith KT, Martin-Brown SA, Florens L, Washburn MP, Workman JL. Chem Biol. 2010;17:65–74. doi: 10.1016/j.chembiol.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balkhi MY, Trivedi AK, Geletu M, Christopeit M, Bohlander SK, Behre HM, Behre G. Oncogene. 2006;25:7041–7058. doi: 10.1038/sj.onc.1209689. [DOI] [PubMed] [Google Scholar]
- 46.Lu Q, Hutchins AE, Doyle CM, Lundblad JR, Kwok RP. J Biol Chem. 2003;278:15727–15734. doi: 10.1074/jbc.M300546200. [DOI] [PubMed] [Google Scholar]
- 47.Gao J, Siddoway B, Huang Q, Xia H. Biochem Biophys Res Commun. 2009;379:1–5. doi: 10.1016/j.bbrc.2008.11.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michael LF, Asahara H, Shulman AI, Kraus WL, Montminy M. Mol Cell Biol. 2000;20:1596–1603. doi: 10.1128/mcb.20.5.1596-1603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henikoff S, Henikoff JG. Proc Natl Acad Sci U S A. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bollen M. Trends Biochem Sci. 2001;26:426–431. doi: 10.1016/s0968-0004(01)01836-9. [DOI] [PubMed] [Google Scholar]
- 51.Leone V, Mansueto G, Pierantoni GM, Tornincasa M, Merolla F, Cerrato A, Santoro M, Grieco M, Scaloni A, Celetti A, Fusco A. Oncogene. 2010;29:4341–4351. doi: 10.1038/onc.2010.179. [DOI] [PubMed] [Google Scholar]
- 52.Canettieri G, Morantte I, Guzman E, Asahara H, Herzig S, Anderson SD, Yates JR, 3rd, Montminy M. Nat Struct Biol. 2003;10:175–181. doi: 10.1038/nsb895. [DOI] [PubMed] [Google Scholar]
- 53.Haider S, Joseph CG, Neidle S, Fierke CA, Fuchter MJ. Bioorg Med Chem Lett. 2011;21:2129–2132. doi: 10.1016/j.bmcl.2011.01.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dowling DP, Gattis SG, Fierke CA, Christianson DW. Biochemistry. 2010;49:5048–5056. doi: 10.1021/bi1005046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dowling DP, Gantt SL, Gattis SG, Fierke CA, Christianson DW. Biochemistry. 2008;47:13554–13563. doi: 10.1021/bi801610c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sengupta N, Seto E. J Cell Biochem. 2004;93:57–67. doi: 10.1002/jcb.20179. [DOI] [PubMed] [Google Scholar]
- 57.Hayakawa T, Nakayama J. J Biomed Biotechnol. 2011;2011:129383. doi: 10.1155/2011/129383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dreiza CM, Brophy CM, Komalavilas P, Furnish EJ, Joshi L, Pallero MA, Murphy-Ullrich JE, von Rechenberg M, Ho YS, Richardson B, Xu N, Zhen Y, Peltier JM, Panitch A. FASEB J. 2005;19:261–263. doi: 10.1096/fj.04-2911fje. [DOI] [PubMed] [Google Scholar]
- 59.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Nat Rev Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee H, Sengupta N, Villagra A, Rezai-Zadeh N, Seto E. Mol Cell Biol. 2006;26:5259–5269. doi: 10.1128/MCB.01971-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koyasu S, Nishida E, Kadowaki T, Matsuzaki F, Iida K, Harada F, Kasuga M, Sakai H, Yahara I. Proc Natl Acad Sci U S A. 1986;83:8054–8058. doi: 10.1073/pnas.83.21.8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taiyab A, Rao Ch M. Biochim Biophys Acta. 2011;1813:213–221. doi: 10.1016/j.bbamcr.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 63.Vannini A, Volpari C, Gallinari P, Jones P, Mattu M, Carfi A, De Francesco R, Steinkuhler C, Di Marco S. EMBO Rep. 2007;8:879–884. doi: 10.1038/sj.embor.7401047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Somoza JR, Skene RJ, Katz BA, Mol C, Ho JD, Jennings AJ, Luong C, Arvai A, Buggy JJ, Chi E, Tang J, Sang BC, Verner E, Wynands R, Leahy EM, Dougan DR, Snell G, Navre M, Knuth MW, Swanson RV, McRee DE, Tari LW. Structure. 2004;12:1325–1334. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 65.Whitehead L, Dobler MR, Radetich B, Zhu Y, Atadja PW, Claiborne T, Grob JE, McRiner A, Pancost MR, Patnaik A, Shao W, Shultz M, Tichkule R, Tommasi RA, Vash B, Wang P, Stams T. Bioorg Med Chem. 2011;19:4626–4634. doi: 10.1016/j.bmc.2011.06.030. [DOI] [PubMed] [Google Scholar]
- 66.Cole KE, Dowling DP, Boone MA, Phillips AJ, Christianson DW. J Am Chem Soc. 2011;133:12474–12477. doi: 10.1021/ja205972n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vannini A, Volpari C, Filocamo G, Casavola EC, Brunetti M, Renzoni D, Chakravarty P, Paolini C, De Francesco R, Gallinari P, Steinkuhler C, Di Marco S. Proc Natl Acad Sci U S A. 2004;101:15064–15069. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nobeli I, Favia AD, Thornton JM. Nat Biotechnol. 2009;27:157–167. doi: 10.1038/nbt1519. [DOI] [PubMed] [Google Scholar]
- 69.Hedstrom L. Chem Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 70.Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, et al. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 71.Weikl TR, von Deuster C. Proteins. 2009;75:104–110. doi: 10.1002/prot.22223. [DOI] [PubMed] [Google Scholar]
- 72.Tsuzuki S, Honda K, Uchimaru T, Mikami M, Tanabe K. J Am Chem Soc. 2002;124:104–112. doi: 10.1021/ja0105212. [DOI] [PubMed] [Google Scholar]
- 73.Sinnokrot MO, Valeev EF, Sherrill CD. J Am Chem Soc. 2002;124:10887–10893. doi: 10.1021/ja025896h. [DOI] [PubMed] [Google Scholar]
- 74.Horovitz A, Serrano L, Avron B, Bycroft M, Fersht AR. J Mol Biol. 1990;216:1031–1044. doi: 10.1016/S0022-2836(99)80018-7. [DOI] [PubMed] [Google Scholar]
- 75.Lombardi PM, Angell HD, Whittington DA, Flynn EF, Rajashankar KR, Christianson DW. Biochemistry. 2011;50:1808–1817. doi: 10.1021/bi101859k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bressi JC, Jennings AJ, Skene R, Wu Y, Melkus R, De Jong R, O'Connell S, Grimshaw CE, Navre M, Gangloff AR. Bioorg Med Chem Lett. 2010;20:3142–3145. doi: 10.1016/j.bmcl.2010.03.091. [DOI] [PubMed] [Google Scholar]
- 77.Bottomley MJ, Lo Surdo P, Di Giovine P, Cirillo A, Scarpelli R, Ferrigno F, Jones P, Neddermann P, De Francesco R, Steinkuhler C, Gallinari P, Carfi A. J Biol Chem. 2008;283:26694–26704. doi: 10.1074/jbc.M803514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schuetz A, Min J, Allali-Hassani A, Schapira M, Shuen M, Loppnau P, Mazitschek R, Kwiatkowski NP, Lewis TA, Maglathin RL, McLean TH, Bochkarev A, Plotnikov AN, Vedadi M, Arrowsmith CH. J Biol Chem. 2008;283:11355–11363. doi: 10.1074/jbc.M707362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 80.Wu R, Wang S, Zhou N, Cao Z, Zhang Y. J Am Chem Soc. 2010;132:9471–9479. doi: 10.1021/ja103932d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kasner SE, Ganz MB. Am J Physiol. 1992;262:F462–467. doi: 10.1152/ajprenal.1992.262.3.F462. [DOI] [PubMed] [Google Scholar]
- 82.Chang CJ, Jaworski J, Nolan EM, Sheng M, Lippard SJ. Proc Natl Acad Sci U S A. 2004;101:1129–1134. doi: 10.1073/pnas.0308079100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bozym RA, Thompson RB, Stoddard AK, Fierke CA. ACS Chem Biol. 2006;1:103–111. doi: 10.1021/cb500043a. [DOI] [PubMed] [Google Scholar]
- 84.Wang D, Hosteen O, Fierke CA. J Inorg Biochem. 2012 doi: 10.1016/j.jinorgbio.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Douglas BE, McDaniel DH, Alexander JJ. Concepts and models of inorganic chemistry. Wiley; New York: 1994. [Google Scholar]
- 86.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang XJ, Gregoire S. Mol Cell Biol. 2005;25:2873–2884. doi: 10.1128/MCB.25.8.2873-2884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haberland M, Mokalled MH, Montgomery RL, Olson EN. Genes Dev. 2009;23:1625–1630. doi: 10.1101/gad.1809209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Garber K. Nat Biotechnol. 2007;25:17–19. doi: 10.1038/nbt0107-17. [DOI] [PubMed] [Google Scholar]
- 90.Montgomery RL, Potthoff MJ, Haberland M, Qi X, Matsuzaki S, Humphries KM, Richardson JA, Bassel-Duby R, Olson EN. J Clin Invest. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tsai SC. Journal of Biological Chemistry. 2002;277:31826–31833. doi: 10.1074/jbc.M204149200. [DOI] [PubMed] [Google Scholar]
- 92.Lee H, Rezai-Zadeh N, Seto E. Molecular and Cellular Biology. 2004;24:765–773. doi: 10.1128/MCB.24.2.765-773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pflum MK, Tong JK, Lane WS, Schreiber SL. J Biol Chem. 2001;276:47733–47741. doi: 10.1074/jbc.M105590200. [DOI] [PubMed] [Google Scholar]
- 94.Wang AH, Kruhlak MJ, Wu J, Bertos NR, Vezmar M, Posner BI, Bazett-Jones DP, Yang XJ. Mol Cell Biol. 2000;20:6904–6912. doi: 10.1128/mcb.20.18.6904-6912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Grozinger CM, Schreiber SL. Proc Natl Acad Sci U S A. 2000;97:7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li X, Song S, Liu Y, Ko SH, Kao HY. J Biol Chem. 2004;279:34201–34208. doi: 10.1074/jbc.M405179200. [DOI] [PubMed] [Google Scholar]
- 97.Kao HY, Verdel A, Tsai CC, Simon C, Juguilon H, Khochbin S. J Biol Chem. 2001;276:47496–47507. doi: 10.1074/jbc.M107631200. [DOI] [PubMed] [Google Scholar]
- 98.Greco TM, Yu F, Guise AJ, Cristea IM. Mol Cell Proteomics. 2011;10:M110 004317. doi: 10.1074/mcp.M110.004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brandl A, Heinzel T, Kramer OH. Biol Cell. 2009;101:193–205. doi: 10.1042/BC20080158. [DOI] [PubMed] [Google Scholar]
- 100.Seto E, Yang X-J. Handbook of Cell Signaling. In: Bradshaw R, Dennis E, editors. Elsevier, Inc. 2010. pp. 2379–2388. [Google Scholar]
- 101.Galasinski SC, Resing KA, Goodrich JA, Ahn NG. J Biol Chem. 2002;277:19618–19626. doi: 10.1074/jbc.M201174200. [DOI] [PubMed] [Google Scholar]
- 102.Cai R, Kwon P, Yan-Neale Y, Sambuccetti L, Fischer D, Cohen D. Biochem Biophys Res Commun. 2001;283:445–453. doi: 10.1006/bbrc.2001.4786. [DOI] [PubMed] [Google Scholar]
- 103.Gupta R, Jung E, Brunak S. In preparation. 2004. [Google Scholar]
- 104.Qiu Y,ZY, Becker M, John S, Parekh BS, Huang S, Hendarwanto A, Martinez ED, Chen Y, Lu H, Adkins NL, Stavreva DA, Wiench M, Georgel PT, Schiltz RL, Hager GL. Mol Cell. 2006;22:669–679. doi: 10.1016/j.molcel.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 105.Pan LN, Lu J, Huang B. Cell Mol Immunol. 2007;4:337–343. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.