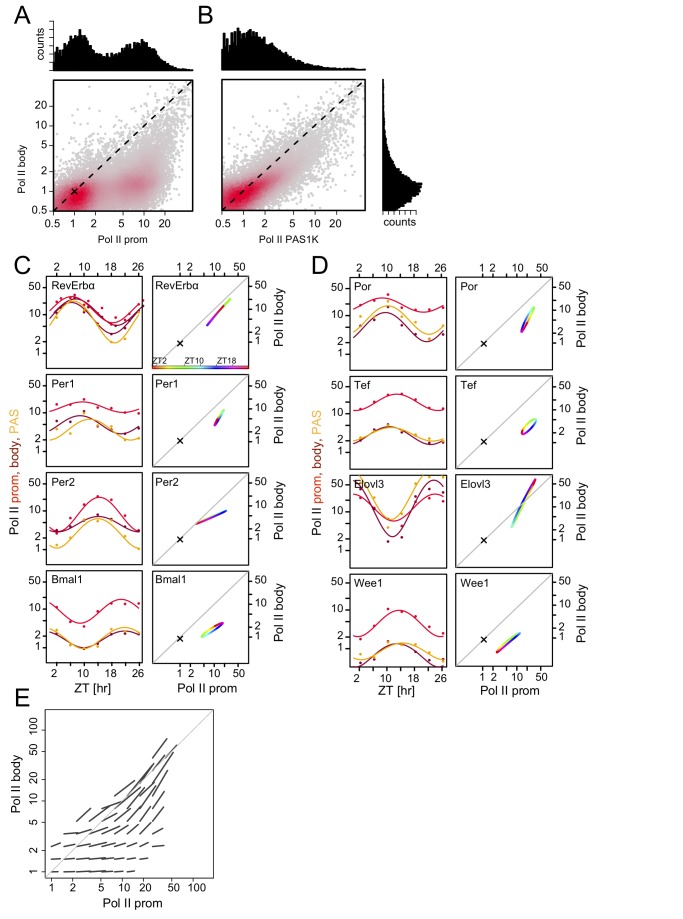
Figure 3. Temporal Pol II occupancies at promoters and in gene bodies oscillate with similar phases.
(A) Pol II occupancy at ZT2 in promoters (mean in ±1 kb regions around TSSs) versus gene bodies (mean over the regions from +1 kb to the PAS) for all genes in logarithmic scale. Color intensity indicates population density. One transcription unit per gene is shown (the selection is based on H3K4me3 and Pol II signals at promoters and PAS, as described in Materials and Methods). Two main populations can be distinguished: one with low Pol II occupancy in both promoter and gene body regions (lower left cloud), corresponding to silent or weakly transcribed genes, and one with higher Pol II occupancy within promoter regions and, to a lower extent, gene body regions (fainter cloud shifted slightly up and to the right). The bimodality of the promoter signal is clearly seen in the projection (histogram above the horizontal axis), whereas the signal in the gene body (elongating polymerases) has a lower dynamic range (histogram on the vertical axis shown in panel B). The cross sign, also shown in panels C and D, indicates background levels estimated from the lower maxima of the histograms. (B) Pol II occupancy at ZT2 in the first 1 kb window after the PAS (PAS1K) versus gene bodies (as in A). The two measures show high correlation, but PAS1K has a larger dynamic range (see Figure 2B). In (A) and (B), data are shown for ZT2, but all time points looked virtually identical. (C and D) Temporal profiles of Pol II promoter/gene body occupancy ratios for core clock genes (C) and selected output genes (D). Left, temporal profiles for promoters (red), gene bodies (brown), and PAS (orange) together with cosine fits. Right, the data from the left panels for the promoter and gene body are plotted against each other in the coordinate system of panel A. ZT times are color-coded (see color bar). Cross signs indicate background levels. Note logarithmic scale on axes. (E) Genome-wide analysis showing that Pol II occupancies at promoters and in gene bodies co-vary in time. Each line shows the average orientation and amplitude of changes during a diurnal cycle for genes in regions on a grid. The nonbinned plot is shown in Figure S3C.