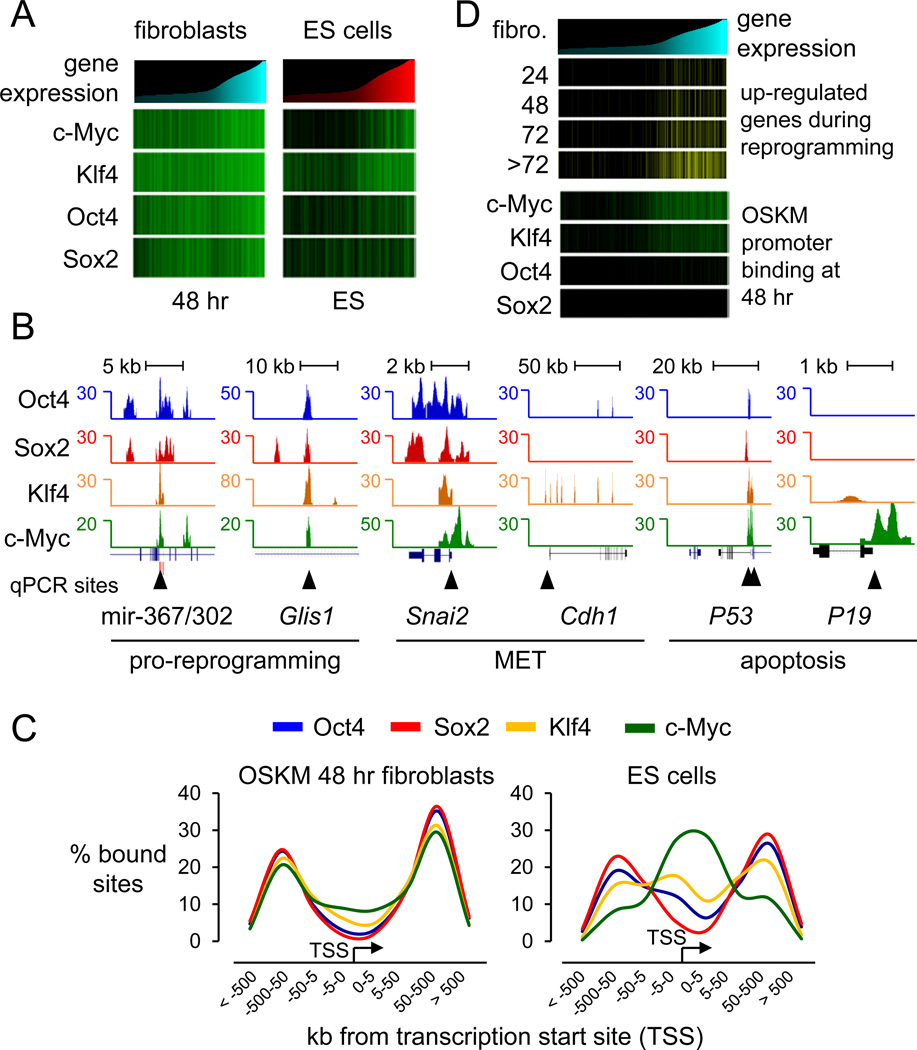
Figure 2. OSKM initially targets genes for reprogramming and apoptosis.
(A) Heatmap analysis showing the correlation between O, S, K, and/or M binding in 48 hr and in ES cells, and gene expression in uninfected human fibroblasts and ES cells (Chen et al., 2008; Lowry et al., 2008; Sridharan et al., 2009). Genes were rank ordered according to their level of expression in fibroblast cells (cyan histogram, left panel) or in ES cells (red histogram, right panel) and correlated with their O, S, K, and/or M occupancy (green llines) at 48 hr (left panel) or in ES cells (right panel), respectively. Table S3 shows expression values in hFib, iPS, and ES cells.
(B) O, S, K, and M ChIP-seq profiles (blue, red, orange, and green respectively) at genes bound at 48 hr and important for reprogramming or apoptosis. OSKM peaks presented are normalized against input DNA sequenced tags. Black triangles indicate regions validated by ChIP-qPCR, as shown in Figure S3C.
(C) O, S, K, and M binding with respect to transcription start sites (TSS) are displayed with the frequency of total binding for Oct4 (blue), Sox2 (red), Klf4 (orange), and c-Myc (green) in 48 hr (left panel) and in ES (right panel, from Chen et al. 2008). See Figure S4 for detailed O, S, K, and/or M binding combination distributions and functional enhancer binding.
(D) Gene expression levels in fibroblasts are rank-ordered from low to high (top cyan histogram). Genes that change expression 2-fold or more over fibroblasts during 24, 48, 72, and >72 hr of reprogramming (Koche et al., 2011) are displayed as yellow lines and correlated with genes occupied by O, S, K, and/or M in their promoters (±1 kb of TSS) at 48 hr (green lines).
See also Figures S2–4.