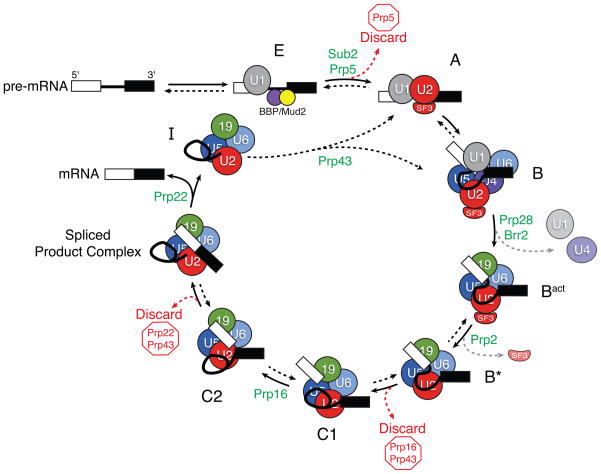
Figure 1.
A model for step-wise spliceosome assembly and catalysis. Specific spliceosomal complexes (E, A, B, and others) are identified according to the human nomenclature. Spliceosome assembly initiates by binding of the U1 snRNP to the 5′ SS and proteins [branch point bridging protein (BBP) and Mud2] to the branch site in E (early) complex. In an ATP-dependent reaction, U2 displaces BBP/Mud2 and binds to the branch site in A complex. B complex then forms by addition of the U4/U6.U5 tri-snRNP. Subsequent to assembly of B complex, catalytic activation requires several additional rearrangements. These include departure of U1 and U4 to form Bact complex (grey arrow); formation of catalytic structures between the pre-mRNA, U2, and U6 snRNAs; and destabilization of several U2 snRNP proteins (the SF3 complex, grey arrow) from the rest of the machinery to form B* complex [8,12,13]. C1 complex is formed after 5′ SS cleavage. For exon ligation, the spliceosome undergoes a conformational change into C2 complex. After the two chemical steps of splicing are complete, the spliceosome enters a disassembly and recycling pathway in which the spliced exons are released and the post-spliceosomal intron product complex (I) is disrupted. Multiple steps in the pathway are promoted by the presence of DExD/H-box ATPases (green). Some of these ATPases have also been implicated in fidelity checkpoints and control the use of discard pathways (red arrows and type) that prevent splicing. As discussed in the text, many of the steps in the pathway have now been shown to be reversible (double arrows), while others have not (single arrows).