Abstract
Reconstructing a functional organ of Corti is the ultimate target towards curing hearing loss. Despite the impressive technical gains made over the last few years, many complications remain ahead for the two main restoration avenues: in vitro transformation of pluripotent cells into hair cell-like cells and adenovirus-mediated gene therapy. Most notably, both approaches require a more complete understanding of the molecular networks that ensure specific cell types form in the correct places to allow proper function of the restored organ of Corti. Important to this understanding are the basic helix-loop-helix (bHLH) transcription factors (TFs) that are highly diverse and serve to increase functional complexity but their evolutionary implementation in the inner ear neurosensory development is less conspicuous. To this end, we review the evolutionary and developmentally dynamic interactions of the three bHLH TFs that have been identified as the main players in neurosensory evolution and development, Neurog1, Neurod1 and Atoh1. These three TFs belong to the neurogenin/atonal family and evolved from a molecular precursor that likely regulated single sensory cell development in the ectoderm of metazoan ancestors but are now also expressed in other parts of the body, including the brain. They interact extensively via intracellular and intercellular cross-regulation to establish the two main neurosensory cell types of the ear, the hair cells and sensory neurons. Furthermore, the level and duration of their expression affect the specification of hair cell subtypes (inner hair cells vs. outer hair cells). We propose that appropriate manipulation of these TFs through their characterized binding sites may offer a solution by itself, or in conjunction with the two other approaches currently pursued by others, to restore the organ of Corti.
Keywords: Inner ear, Development, Hair cell, Restoration, Transcription factor
Introduction
Hearing loss of various forms afflicts over 200 million people, including half of individuals over the age of 65, making sensorineural hearing loss one of the most frequent neurosensory disorders worldwide. Many forms of severe sensorineural hearing loss result from the progressive and near-complete loss of all hair cells in the organ of Corti (OC). A cochlear implant, the only treatment currently at hand, incompletely restores hearing by providing a limited set of stimulation points to the remaining spiral ganglion cell processes, which may be lost over time in humans as they are in animal models (Alam et al. 2007; Shibata et al. 2010). While a cochlear implant is, for a growing number of elderly patients, the only way to restore at least some level of communication ability, the functional OC is clearly “the world's best hearing aid” (Puligilla and Kelley 2009). Therefore, numerous attempts to fully restore hearing are currently centered on two principal approaches:
-
A)
In vitro transformation of embryonic or induced stem cells into hair cell-like cells (Kopecky and Fritzsch 2011; Oshima et al. 2010; Ronaghi et al. 2012), followed by seeding them into the hair cell-depleted cochlea to differentiate as hair cells.
-
B)
In vivo manipulation of proliferation of adult ear cells to increase the number of supporting cells, followed by converting them into hair cells through regulated differentiation with genes inserted into adenoviral vectors (Batts et al. 2009; Brigande and Heller 2009; Izumikawa et al. 2005, 2008; Kopecky and Fritzsch 2011).
Impressive progress in these two areas elevates hair cell regeneration into the realm of possibility as far as generating in vitro hair cell-like cells (Oshima et al. 2010) or transforming in vivo supporting cells into hair cells (Izumikawa et al. 2005, 2008). Both approaches are in line with the state of the art in other systems where simple replacements, such as the trachea coated with induced stem cells of a given patient, are possible (Jungebluth et al. 2011; Jungebluth and Macchiarini 2011). However, in contrast to other systems, the more complex OC has a highly ordered distribution of two different subtypes of hair cells (inner hair cells, IHCs; outer hair cells, OHCs) surrounded by seven different subtypes of supporting cells. This sophisticated organization is functionally meaningful for the perception of sound ranging from 2 to 20 kHz in human and to over 70 kHz in mouse. While restoring only IHCs may provide improvement over no hair cells, restoration of both subtypes of hair cells in specific positions is likely necessary to restore normal hearing. Simply speaking, specific subtypes of hair cells have to be regenerated in a highly stereotyped pattern to ensure function of the restored OC. To achieve such topologically restricted cell type differentiation requires recapitulating the differentiation process during development in therapeutic regeneration.
Multiple developmental studies have made it clear that the transformation of the naïve otic placode epithelium into the neurosensory components of the ear is a stepwise process (Ahmed et al. 2012a; Nichols et al. 2008; Ohyama and Groves 2004; Zou et al. 2008). At the end of this process, topographically restricted expression of transcription factors (TFs) “prime” the otic epithelium to respond to locally expressed diffusible factors such as Fgfs and Bmps to develop the sophisticated microanatomy of the OC (Dabdoub et al. 2008; Fritzsch et al. 2011; Ohyama et al. 2011; Puligilla and Kelley 2009). Existing evidence indicates that the specification of hair cell subtypes depends on certain gene regulations as mutants have been described that specifically lose most IHCs or most OHCs (Ahmed et al. 2012a; Brooker et al. 2006; Deol 1981; Holley et al. 2010; Huh et al. 2012; Pan et al. 2012), though the molecular mechanism remains unclear. Furthermore, we have just begun to understand the emerging feedback loops between different diffusible factors (Huh et al. 2012) and TFs within (Jahan et al. 2012; Pan et al. 2012) and between cells of the OC (Basch et al. 2011) and their roles in regulating cell differentiation. Obviously, a much deeper understanding of the sequential activation of genes and the interactions between them during development of the OC is needed before we can attempt to recapitulate the formation of specific hair cell subtypes in specific places with either of the two above-indicated approaches.
Studies in the mouse suggest that the expression of many differentiation factors is progressively downregulated in aging animals, in particular, following disruption of the OC development (Groves 2010; Pan et al. 2011, 2012), leaving only a small number of those factors to be permanently expressed, such as Foxg1 (Pauley et al. 2006) and Gata3 (Duncan et al. 2011; Karis et al. 2001). It is therefore possible that the molecular guidance for topologically correct differentiation of hair cells is insufficient in the adult cochlea, making cell-specific targeting seemingly unresolvable at our current level of understanding. Indeed, treatment with TFs that effectively regenerate hair cells in embryos cannot achieve the same effect in the adult cochlea devoid of an OC (Izumikawa et al. 2008). However, in analogy to the TFs and microRNAs needed to reprogram cells to form inducible pluripotent stem cells (Rosa and Brivanlou 2011), it may be possible to upregulate a limited set of TFs and microRNAs (Ahmed et al. 2012a; Soukup et al. 2009) to “prime” the epithelium to respond with differentiation upon expression of cell type-specific TFs such as Atoh1, something the adult “flat epithelium” is incapable of doing on its own (Izumikawa et al. 2008). Precisely which TFs and ear specific microRNAs and other factors are necessary for “priming” remains to be fully elucidated.
Alternatively, a third approach to restore a functional OC is to directly transform existing non-sensory epithelial cells of the “flat epithelium” (Izumikawa et al. 2008; Pan et al. 2011) into a new OC by activating the necessary set of TFs and other factors (Ahmed et al. 2012a; Fritzsch et al. 2011). As a proof of principle, recent data suggest that direct transformation of skin-derived cells into neurons is possible (Lujan et al. 2012; Pang et al. 2011) and neuron-specific microRNAs can transform fibroblasts into neurons (Yoo et al. 2011). Obviously, this approach is at the moment still far removed from translation into restoration of the OC but could embody the ultimate solution. Again, molecular dissection of the interactions and cross-regulation and a reasonable understanding of transcriptional regulations of the critical TFs and microRNAs for OC development are required for the elucidation of the right combination of factors needed to accomplish this goal.
Ultimately, no matter where we start, the task remains the same: ensuring that specific subtypes of hair cells develop in specific positions and drive topologically correct differentiation of supporting cells to restore a functional OC. Unfortunately, defining these specific cell types and subtypes seems to depend on a growing set of TFs and diffusible morphogens (Ahmed et al. 2012a; Basch et al. 2011; Fritzsch et al. 2011; Groves and Fekete 2012; Huh et al. 2012; Ohyama et al. 2011) with as yet mostly unclear interactions and hierarchies. Below, we will review the best-characterized TFs that execute the neurosensory differentiation in the ear, the basic helix-loop-helix (bHLH) TFs and their roles in cell type specific differentiation. We will introduce the evolution of these factors and how bHLH TF evolution ties into hair cell evolution. This analysis will be followed by an assessment of the TFs' molecularly dissected functions to direct the development of specific hair cell subtypes of the OC. We will then explore the possibility to regulate the expression of genes downstream to these factors using the emerging knowledge of the binding of these TFs to specific promoter regions of molecularly distinct E-boxes. Finally, we will provide a novel perspective on how to use recently generated complex mutant mice to understand the molecular tuning of specific cell types independent of the topological information.
The evolution of bHLH proteins predates the evolution of inner ear neurosensory cells
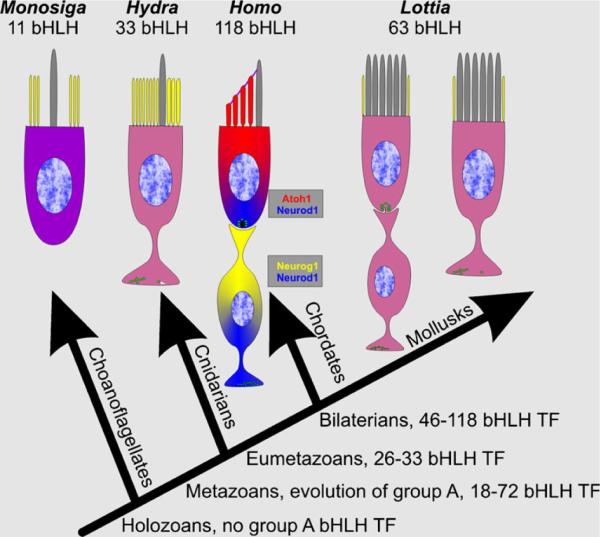
bHLH TFs belong to ancestral pro-metazoan TFs that are already found in single-celled ancestors of metazoans such as fungi and choanoflagellates, the latter being the likely ancestor of metazoans (Degnan et al. 2009; Gazave et al. 2009; Young et al. 2011). What role the bHLH TFs played in the single-celled metazoan ancestors remains speculative but the function of some bHLH genes such as the Myc genes appears highly conserved from yeast to the mammalian ear (Kopecky et al. 2011; Schuldiner et al. 1996; Young et al. 2011). Other conserved metazoan bHLH TFs such as Twist1 mediate transitions from proliferation to differentiation, including migration (Lee and Yutzey 2011). Such variable function from proliferation to differentiation is also found in proneural bHLH TFs (Ali et al. 2011). Among metazoans, bHLH genes have diversified by an order of magnitude (from about 10 in choanoflagellates to 118 in humans) but the small number of bHLH TFs in choanoflagellates could also represent a secondary reduction (Sebe-Pedros et al. 2011).
It appears that a specific group of bHLH TFs, the group A bHLH TFs (Simionato et al. 2008) that contain factors essential for cell fate specification, evolved together with a range of other TFs in metazoan ancestors (Galliot and Quiquand 2011). Several members of these group A bHLH TFs such as Atoh1, Neurog1 and Neurod1, as well as members of other TF families such as Foxg1 (Pauley et al. 2006), Pax2/5/8 (Bouchard et al. 2010), Pou4f3 (Xiang et al. 2003), Lmx1a (Nichols et al. 2008) and Eya1/Six1 (Ahmed et al. 2012a; Ahmed et al. 2012b; Zou et al. 2008), are now known to be essential for the ear or other organ development. Furthermore, the diversification of bHLH TFs was accompanied by a diversification of their E-box DNA binding sites (between 600 and 1,000 partially overlapping genes; Klisch et al. 2011; Seo et al. 2007) and an increase in the number of cell types (from 1 to over 200 distinct cell types recognized in humans). In essence, the correlated changes in DNA binding properties and DNA binding sites resulted in co-evolution and diversification of the roles of the bHLH TFs and their binding sites (Degnan et al. 2009; Yang et al. 2011).
In single-celled organisms, the 10–31 bHLH TFs identified seem to play a role in regulating proliferation and possibly in transitioning from one phase of the life cycle into another, such as the shift between mitosis and growth (Sebe-Pedros et al. 2011). The metazoan group A bHLH TFs play an apparently similar role in the developing cortex (Ali et al. 2011), cerebellum (Klisch et al. 2011), cochlear nuclei (Maricich et al. 2009) and intestine (Li et al. 2011; Shroyer et al. 2007). Unlike non-group A bHLH TFs, which have retained and are mostly limited to the function of regulating mitosis (Young et al. 2011), group A bHLH TFs evolved the additional function to regulate topographically distinct cell differentiation in metazoans. It is mostly unclear how exactly the topographically restricted expression of these factors in only a subset of precursors is achieved but numerous mutant analyses have demonstrated that restricted expression of bHLH TFs provides a crucial developmental step that cannot be replaced by other TFs. It is possible that the large number of downstream genes regulated by each of these factors create non-redundant pathways (Klisch et al. 2011; Seo et al. 2007). However, this cannot be the only reason for bHLH diversification as some other TFs such as the Pax genes regulate even larger numbers of downstream genes and are typically not recruited for cell fate but rather organ fate determination (Bouchard et al. 2010; Kozmik et al. 2003; O'Brien and Degnan 2003). Nonetheless, it is likely that the group A bHLH TF family evolved into their diverse assortment of cell fate determination roles because of the ancestral function of bHLH genes in regulating various stages of a cell's life cycle. Indeed, in most metazoans, there is a life-long balance between cell cycle and cell differentiation, with the mammalian ear and central nervous system being on the extreme end of differentiation without any overt ability to regenerate hair cells and neurons via reinitiating proliferation. In this context, it is noteworthy that the function of one group A factor, atonal/Atoh1, can be regulating proliferation (Klisch et al. 2011), differentiation (Bermingham et al. 1999), or both (Fritzsch et al. 2010; Shroyer et al. 2007).
If this idea is correct, we would need to study in more detail the expression of different bHLH genes of the same group or of different groups, using modern RT-qPCR approaches to monitor expression changes over time (Brar et al. 2011) in very short intervals in the same cells as a prerequisite towards evaluating the complexity of cross-regulation. This knowledge would be necessary to understand why in certain species of vertebrates all precursors are depleted through differentiation, while, in other cases, precursors are set aside as adult stem cells to repopulate lost cells when needed or even continue to proliferate throughout life.
bHLH transcription factors that are involved in proliferation in the ear
In the ear, two bHLH proto-oncogenes, N-Myc and L-Myc, are mostly co-expressed (Kopecky et al. 2011), whereas a third gene of the same family, C-Myc, seems to play little if any role (Dominguez-Frutos et al. 2011). Conditional deletion of N-Myc in the ear shows a massive reduction of ear growth, apparently through reduced proliferation (Kopecky et al. 2011). Consistently, overexpression of N-Myc shows increased ear size through increased proliferation (Dominguez-Frutos et al. 2011). However, N-Myc is not only expressed in the developing ear but is also expressed later on in differentiated hair cells. It is puzzling that a proto-oncogene is expressed in terminally differentiated cells that are unable to undergo induced mitosis but will die through apoptosis if so induced (Mantela et al. 2005). At a molecular level, N-Myc could alter the bHLH signaling through interactions with ID proteins and/or alter the microRNA content of differentiating hair cells (Kopecky and Fritzsch 2011; Kopecky et al. 2011) possibly through interactions with other TFs such as Eya1-Six1 (Ahmed et al. 2012a). Notably, the initially differentiating hair cells of the OC but not the vestibular system, eventually die in the N-Myc conditional knockout (CKO) mutant, indicating that N-Myc plays a yet to be defined unique role in the OC hair cells. Early expression studies of bHLH genes associated with neurosensory differentiation in the ear (Neurog1, Neurod1, Atoh1, Nhlh1, Nhlh2) in the absence or presence of N-Myc will help to elucidate its additional function in regulating ear neurosensory development. Moreover, comparable work on L-Myc CKO mutants is needed to verify the possible functional redundancy of these two proto-oncogenes.
Understanding the molecular regulation of cell cycle control and the balance between proliferation and differentiation is essential for the attempts to restore hearing. For restoration along avenue “A”, which requires the initial formation of stem cells followed by seeding these cells into their topologically restricted positions along the OC, it is necessary to precisely control the cell cycle to not only acquire the correct number of physiologically normal hair cells but also to be able to inhibit extended proliferation when cell numbers are met and differentiation must occur (Jeon et al. 2011; Kopecky and Fritzsch 2011). Uncontrolled proliferation would create a cancer-like state whereas a lack of profound proliferation would create too few cells that may eventually die, as seen in the N-Myc CKO mice (Kopecky et al. 2011). The task may be even more daunting for restoration avenue “B”, as the in vivo post-mitotic cells to be manipulated have long since exited the cell cycle and are not normally responsive to proliferation cues, unlike the stem cells in avenue “A”. No matter the starting point, in forcing cell cycle re-entry in differentiated cells, not only must the correct number of cells be formed by a controlled balance between proliferation and differentiation as in avenue “A” but this re-entry into the cell cycle must produce the normal cytoarchitecture of the OC. Nonetheless, re-entry through inhibition of cell cycle inhibitors (Laine et al. 2007; Liu and Zuo 2008; Minoda et al. 2007; Oesterle et al. 2011; Ono et al. 2009; Rocha-Sanchez et al. 2011; Sulg et al. 2010; Weber et al. 2008; Yu et al. 2010) or over-expression of proto-oncogenes (Loponen et al. 2011; Ozeki et al. 2007) have been successful for at least a short period of time but all newly formed cells eventually undergo apoptosis as there appears to be a more complicated intrinsic regulation than previously thought (Huang et al. 2011). Because of the possibility of cross-talk between bHLH TFs and the intrinsic interplay evident with the Mycs, IDs and differentiation cues such as Atoh1 and Neurod1, the understanding of both the positive (proto-oncogenes such as N-Myc) and negative (tumor suppressors such as p21/27Kip1 and pRb) regulation of the cell cycle has never been more important to hair cell restoration. Ultimately, to fully control the cell cycle, it is likely that manipulation of protooncogenes, tumor suppressors and differentiation TFs must be fully utilized, just as the gas and brakes must be in a car; otherwise, the cell cycle will speed out of control or stop dead in its tracks.
bHLH transcription factors that are involved in differentiation in the ear
Mutational analysis in mice has identified three closely related group A bHLH TFs (Atoh1, Neurog1, Neurod1) that are essential for neurosensory cell differentiation (Bermingham et al. 1999; Kim et al. 2001; Ma et al. 2000). Based on data on replacement of the mouse Atoh1 by the fly atonal gene showing that they can fully substitute each other for normal hair cell differentiation (Wang et al. 2002), it was assumed that atonal might be close to the ancestral bilaterian gene and thus it has served as the founder for a growing family of atonal-like genes. atonal-like bHLH genes have been found in diploplastic metazoans (Seipel et al. 2004), which have mechanosensory cells with a high degree of similarity to choanocytes of sponges and choanoflagellates (Burighel et al. 2011; Fritzsch et al. 2007; Jorgensen 1989). In these animals, the sensory cells always have their own axon (Fig. 1) that feeds sensory information into the nerve net for appropriate responses. In contrast, derived bilaterians such as vertebrates or cephalopods have sensory cells without an axon and sensory neurons connecting these sensory cells to the central nervous system (Fritzsch et al. 2010). From this observation, it follows that some bilaterians have evolved molecular means to convert a single neurosensory cell type into two distinct cell types. We have proposed that the multiplication of bHLH genes is tied into this cellular diversification (Fritzsch et al. 2000), although the exact triggering mechanism remains speculative.
Fig. 1.
Available data relevant for the evolution of human hair cells and sensory neurons are shown. Note that Monosiga has 11 bHLH TFs, none of which are orthologous to Metazoans. Eumetazoans have sensory cells with axons and display asymmetric distribution of microvilli (yellow) and kinocilium (gray). In mammals, the three bHLH TFs are partially overlapping to drive neuronal (Neurog1, Neurod1) and hair cell (Atoh1, Neurod1) development. A superficially similar arrangement is found in some mollusks but which bHLH genes are expressed in these cells is unknown. Compiled after Budelmann (1992), Degnan et al. (2009), Fritzsch et al. (2007) and Galliot and Quiquand (2011)
It was proposed that the apparently newer bHLH genes of the atonal family that play a role in neuronal development, Neurod1 and Neurog1, have evolved later (Simionato et al. 2007), as they were specifically associated with the neuronal precursors. However, more recent data, including multiple genome sequences of basic metazoans, show that the situation in flies is uniquely derived through the loss of Neurod1 (Simionato et al. 2008). Most importantly, using a non-atonal-related bHLH gene as an out-group, a maximum likelihood tree suggests that Neurog1 is closer to the atonal family root, with Neurod1 being more derived and atonal/Atoh1 being most derived (Simionato et al. 2008). Obviously, if more data confirm this reformed evolutionary relationship, it appears that the sequence of discovery of these genes in flies may have biased our perception of their evolution and the atonal family of genes should be renamed the neurogenin family of genes.
No matter the ancestry of these bHLH TFs, the multiplication of these closely related bHLH genes happened in basal bilaterians and resulted in an initial co-expression of these three TFs in the same cell. As indicated in numerous studies on such gene duplication, there is a limited chance that each gene evolved into a novel regulatory cascade by activating a restricted set of downstream genes through evolutionary changes in their DNA binding sites. Essentially, TFs and their DNA binding sites co-evolve (Yang et al. 2011), much like the better studied co-evolution of hormones and their receptors (Bridgham et al. 2006). Presumably, the bHLH gene multiplication generated a set of redundant signaling molecules that provided the robust basis needed to explore novel functions without compromising the essential basic function (Espinosa-Soto et al. 2011; Wagner 2011). Ultimately, a random mutational walk through gene-space via mutagenesis (Wagner 2011) may have resulted in the specific association of each TF with a specific aspect of cellular evolution: a split of the simple precursor that generated only one neurosensory cell type into two partially overlapping precursor populations was accomplished in vertebrate ancestors. This partial split resulted, through an as yet only partially explored interactive developmental cascade (see below), in the differentiation of two distinct cell types, the sensory hair cell and the sensory neuron of the vertebrate ear (Fritzsch et al. 2006, 2010).
Many metazoans have these three bHLH genes; however, only some species have, most possibly convergently, explored the innovatibility inherent in such gene multiplication (Wagner 2011) and evolved a morphologically dissimilar set of sensory cells connected by neurons to the central nervous system (Budelmann 1992; Burighel et al. 2011). Unfortunately, no expression or experimental data on bHLH genes exist in these specific non-vertebrate animals, making it currently impossible to establish molecular similarities or differences in how these animals arrived at a comparable cellular diversification by evolving the bHLH genes into a developmental module that can generate two distinct cell types instead of one. In particular, cephalopods, with the co-existence of sensory cells with their own axon next to sensory cells that are connected to the central nervous system via sensory neurons (Budelmann 1992), need to be analyzed to understand how bHLH genes are regulated into such distinct outcomes. It is notable that flies have apparently lost one of those three ancestral genes (Neurod1) but have also evolved novel multiplications of bHLH genes that seem not be tied into the generation of radically different cell types but rather in the modification of existing types (Cachero et al. 2011). It is conceivable that expression of these bHLH genes in the brain or intestine could have driven their evolution and their implementation in the evolving neurosensory cell types was secondary. However, the basic evolutionary principle of gene duplication followed by cell type multiplication, further outlined below, would remain the same.
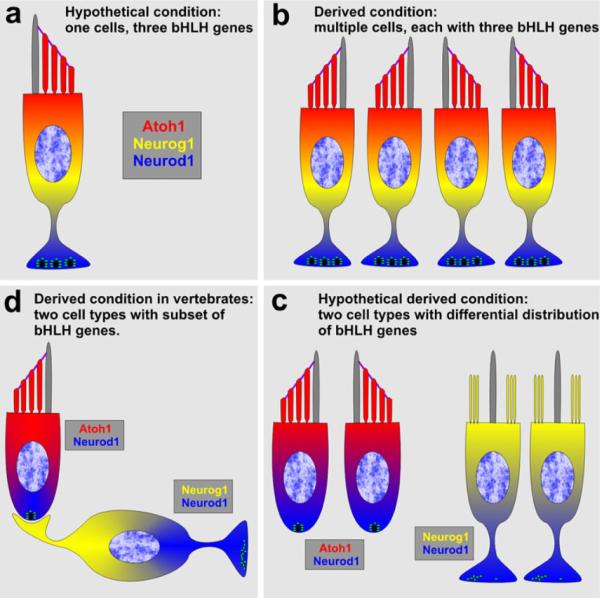
Once a sensory cell with an axon had evolved the expression of three bHLH TFs, the precursors for this single cell were multiplied to form an array of sensory cells through an enhanced proliferation of a precursor population, instead of a single cell (Fig. 2a, b). Such an array adopted the universal planar cell polarity signal to adjust the sensory cells within such an epithelium into a distinct and typically opposing, polarity as found in virtually all mechanosensory epithelia (Fig. 2b). Overlapping with this multiplication, a segregation of bHLH gene expression must have occurred and the increased populations were reorganized into two partially segregated populations in terms of bHLH TF expression. Discrete differences in overlap of diffusible factors combined with the evolutionary changes in bHLH gene activation patterns likely evolved as a consequence of the increased size and limitations in diffusible gradients as well as the multiplication of these diffusible factors; for example, the Fgf, Wnt, Bmp and HH families (Groves and Fekete 2012). As a consequence, some cells at one end of the larger array of uncommitted cells could have developed as sensory neurons without mechanotransduction abilities, whereas cells at the other end of the array could have developed as hair cells without an axon but with mechanotransduction capabilities (Fig. 2c). These different developmental trajectories evolved as a consequence of the gene multiplication, mutations in their respective promoter regions and consequently, a slight difference in response to diffusible factors. Once such a segregated developmental pathway had evolved, the pairing of neuronal and hair cell development was fixed permanently, forming a novel pathway to ensure that specific hair cells and associated sensory neurons had to evolve to guarantee an epithelial- and polarity-specific presentation stimuli in the central nervous system (Fig. 2d). In the second part of this review, we will provide developmental data that are consistent with such an evolutionary scenario and show that many neurons derive from one part of a sensory epithelium or from areas between sensory epithelia (Fig. 3) (Fritzsch et al. 2002).
Fig. 2.
A hypothetical evolutionary transformation of a single sensory cell with an axon expressing three bHLH TFs (a) into two connected hair cell/sensory neuron (d). Hypothetical intermediate steps could be the formation of a multicellular array (b) that allowed differential expression of the three bHLH TFs to induce formation of two distinct cell types (c). Once these two cell types were formed, additional changes led to the two cell types recognized in vertebrate ears (d)
Fig. 3.
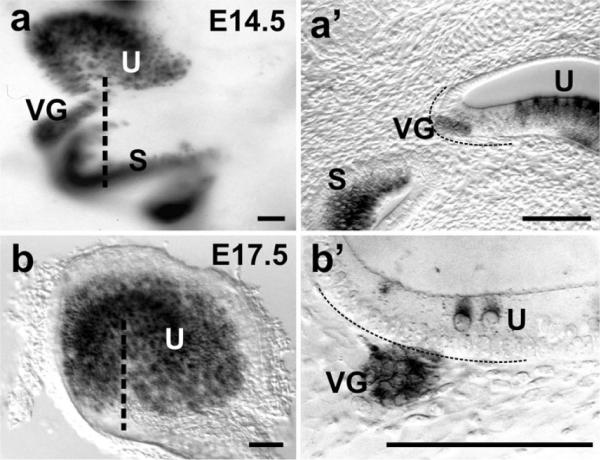
This image shows the delamination of Bdnf–LacZ positive neurons from two sensory epithelia, the utricle (U) and saccule (S). Note that the area of delamination of neurons is devoid of hair cell labeling at E14.5 in the whole-mounted ear (a). Thick sections at the plane indicated in (a) by a dotted line show that vestibular ganglion neurons (VG) are at the neuronal edge of the utricle and separated by a region of proliferating precursors that are not yet differentiating as either neurons or hair cells (a'). At E17.5, only a few neurons remain immediately underneath the utricle and are clearly distinct from the differentiated hair cells (b'). S saccule, U utricle, VG vestibular ganglion. Bar 100 μm
A critical step in this scenario is the multiplication of sensory cell precursors into a larger array that allowed differential expression of bHLH genes as a consequence of relaxed expression specification within that array through differential signal strength of diffusible factors (Groves and Fekete 2012). Both overlapping of multiple bHLH genes as well as multiplication of a single precursor are prerequisites for the innovation to generate a novel cell type: the sensory neuron without mechanotransduction through relaxed, altered cell fate specification.
In summary, in basal metazoans an ancestral bHLH TF that was originally involved in regulating the development of the highly conserved mechanosensory cell morphology (Burighel et al. 2011; Fritzsch et al. 2007) was multiplied (Fritzsch et al. 2010; Simionato et al. 2008). This multiplication provided the genetic robustness needed for the innovative split of the ancestral sensory cell with its own axon into two cells: a hair cell that specializes in mechanotransduction and a sensory neuron that connects the hair cell to the central nervous system (Fig. 1). Below, we will highlight the complexity of the TF interaction in mice during development and speculate how it could have been tied into the evolutionary split of a simple precursor population into two partially overlapping populations that generate the two discrete cell types, as well as the two subtypes of the hair cells.
bHLH transcription factors regulate both type- and subtype-specific development of inner ear neurosensory cells
In principle, cell fate decision making in any developing system (including the ear) follows three steps (Lander 2011; Peter and Davidson 2011):
-
1)
Diffusible signals establish, through gradients, the position of specific cell types;
-
2)
Regulation of various factors “prime” cells to respond appropriately to other topologically more specific TFs with cell type-specific differentiation;
-
3)
Consolidation of cell fate through cell–cell interactions ensures a coordinated assembly of multiple cell types in a functionally relevant pattern.
This general principle applies to the OC development and uses the following known factors for each level of molecular regulation:
-
1)
Bmps, Fgfs, Shh and Wnts are diffusible factors that define the otic placode and later sensory epithelia (Bok et al. 2007; Chang et al. 2008; Groves and Fekete 2012; Huh et al. 2012; Ohyama et al. 2011; Pauley et al. 2003; Pirvola et al. 2000; Riccomagno et al. 2002; Riccomagno et al. 2005).
-
2)
Eya1, Six1, Foxg1, Gata3, Jag1, Pax2, Sox2 and other genes act as cell fate primers to solidify the transient pattern generated by the diffusible factors (Ahmed et al. 2012a; Bouchard et al. 2010; Fritzsch et al. 2006, 2011; Kiernan et al. 2006; Pauley et al. 2006; Zou et al. 2008).
-
3)
At least three proneural bHLH TFs (Atoh1, Neurod1, Neurog1) define and differentiate sensory neurons and hair cells (Bermingham et al. 1999; Kim et al. 2001; Ma et al. 1998, 2000). In addition, neurogenic TFs regulate the initial patterning of supporting cells and the overall pattern of the OC development, through both the Delta/Notch system and through feedback loops of diffusible factors, such as Fgf8 expressed in IHCs (Basch et al. 2011; Doetzlhofer et al. 2009; Fritzsch et al. 2011; Jacques et al. 2007; Pirvola et al. 2000).
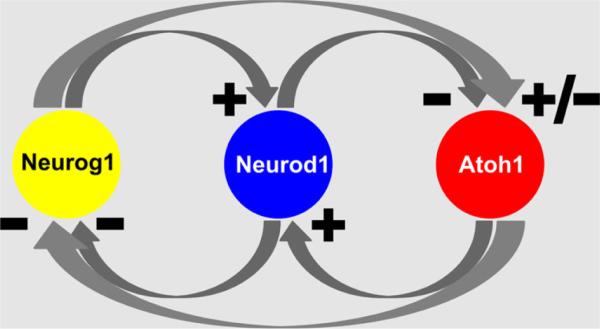
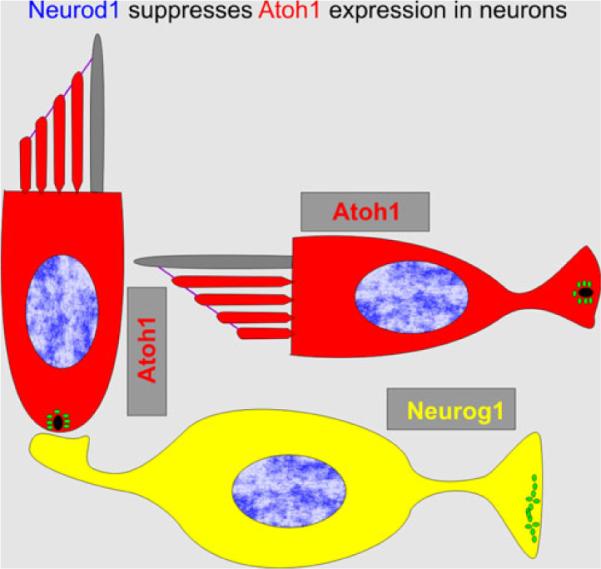
Although original loss-of-function studies show a simple one gene–one cell type developmental scheme, the results of various more detailed knockout studies and expression analysis show that the three proneural bHLH TFs regulate neurosensory development through extensive intra- and intercellular interactions (Fig. 4). For example, Neurog1 drives the expression of Neurod1 in sensory neurons (Ma et al. 1998) and deletion of either gene leads to neuronal loss. Deletion of Neurog1 also results in hair cell loss and premature expression of Atoh1 (Ma et al. 2000; Matei et al. 2005). This suggests that Neurog1 inhibits Atoh1, which in turn cross-regulates expression of Neurog1 (Jahan et al. 2012; Matei et al. 2005; Raft et al. 2007). In addition, Atoh1 expression is now known to be necessary for Neurod1 expression in developing hair cells (Jahan et al. 2012; Pan et al. 2012). Thus, possibly through disinhibition of Atoh1 expression, absence of Neurog1 increases expression of Neurod1 in hair cells (Matei et al. 2005). Neurod1 also suppresses both Neurog1 (Jahan et al. 2010b) and Atoh1 expression (Jahan et al. 2010b). These data indicate an unprecedented level of complexity of the cross-regulation of these three bHLH TFs with a developmentally incomplete segregation of expression and interaction (Fig. 4). We hypothesize that this incomplete developmental segregation is possibly a consequence of the incomplete evolutionary segregation of their initially identical developmental signaling of both downstream and upstream expression regulation. In essence, while at least some hair cells will differentiate in the absence of either Neurog1 or Neurod1, the presence of both Neurog1 and Neurod1 is necessary to balance the proportion of neurons to hair cells.
Fig. 4.
Known intra- and intercellular interactions of Atoh1 bHLH TF family members are depicted. Neurog1 and Atoh1 are necessary for Neurod1 expression in neuronal and hair cell precursors, respectively. Neurod1, in contrast, inhibits Neurog1 and Atoh1 in neurons and hair cells, respectively. Possible intercellular interactions cross-regulate Atoh1/Neurog1 expression. Atoh1 appears to inhibit Neurog1, whereas absence of Neurog1 results in disinhibition of Atoh1. Neurog1 can reduce hair cell development in some parts of the ear but also results in extra hair cells in other parts but it is unclear how this relates to cross-regulation of Atoh1
Beyond the cell decision-making process to differentiate sensory neurons, hair cells and supporting cells, the OC development also requires the specification of morphologically and physiologically distinct subtypes of hair cells (IHCs and OHCs), sensory neurons (type I and II spiral ganglion neurons) and supporting cells (seven subtypes) at specific positions for proper function. It is unclear how these different cell subtypes form in their specific positions and what unique mixture of TFs drives their differentiation. As progress in hair cell regeneration is inching toward hair cell restoration, the as yet unspecified pathways and molecular cues to make different subtypes of hair cells in specific locations will become crucial for the reconstitution of a functional OC (Yang et al. 2012). To achieve reconstruction, we will need to understand how to generate IHCs at the correct position as generation of OHCs or vestibular hair cells instead might not provide the proper mechanoelectric sound transduction needed for hearing. Below is a summary of our recent studies on the cell fate decision-making in the ear.
Formation of neurons and sensory hair cells results from interplay of bHLH TFs
Most revealing for a more detailed understanding of the intracellular cross-regulation of these bHLH factors are experiments that remove Neurod1. Neurod1 loss results in degeneration of most of the inner ear sensory neurons (Jahan et al. 2010a; Kim et al. 2001). Since some of the other bHLH TFs in the developing ear have the capacity to rescue some neurons (Kruger et al. 2006) but apparently not to suppress Atoh1, the de-repressed Atoh1 expression converts some neurons into hair cells through the maintenance of the initial expression of Atoh1 (Figs. 5, 6). Selective overexpression of Atoh1 in the developing sensory neurons is now needed to show how widespread this effect of transformation into hair cells can be in sensory neurons and how much the Neurog1 mediated expression of Neurod1 in neurons is not only needed in normal development but can counteract an induced Atoh1 expression.
Fig. 5.
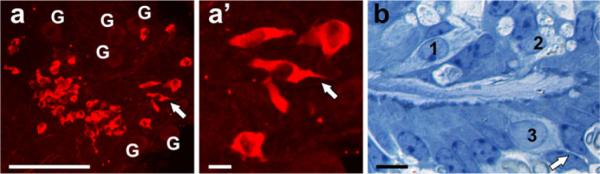
Deletion of Neurod1 results in loss of many sensory neurons. Among the surviving neurons some develop as quasi-hair cells through expression of Atoh1 and Myo7a, two well-accepted markers for hair cells in the ear. Occasionally, these hair cells develop short processes that may contain the synaptic contacts, as indicated. For details, see (Jahan et al. 2010b; Sebe-Pedros et al. 2011)
Fig. 6.
Deletion of Neurod1 results in transformation of cells, most likely sensory neurons into Myo7a positive “hair cells” (a). These cells are typically irregular or spindle-shaped and some of them display a more or less extended projection on the opposite part of the stereocilia/kinocilia, indicating a distinct polarity of the apex and the base of hair cells (arrows in a and a'). Most of these Myo7a-positive cells are found along ventricles formed inside the ganglia, into which they extend with their apical specializations. While some of these cells display multiple vesiculated structures near their base (1, 2 in b), others display a basal extension somewhat similar to the axon emanating from a neuron (3, arrow in b). G ganglion neurons. Bars (a) 100 μm, (a', b) 10 μm
Equally interesting is our data on a targeted misexpression of Neurog1 under the Atoh1 promoter control. Based on the fact that absence of Neurod1 alters hair cell subtype differentiation as it results in a higher and earlier expression of Atoh1 (Jahan et al. 2010b), one would expect that co-expression of one allele of Atoh1 and one expressing Neurog1 would alter the hair cell phenotype due to additional disabling of the Atoh1 signaling. Indeed, that is what we recently reported in a Atoh1kiNeurog1 knockin mouse model (Figs. 7, 8) (Jahan et al. 2012). However, while Atoh1 upregulation in sensory neurons, after eliminating the repressor Neurod1, leads to hair cell differentiation, no differentiation of hair cells as neurons can be induced with homozygotic misexpression of Neurog1 under the Atoh1 promoter control (Jahan et al. 2012).
Fig. 7.
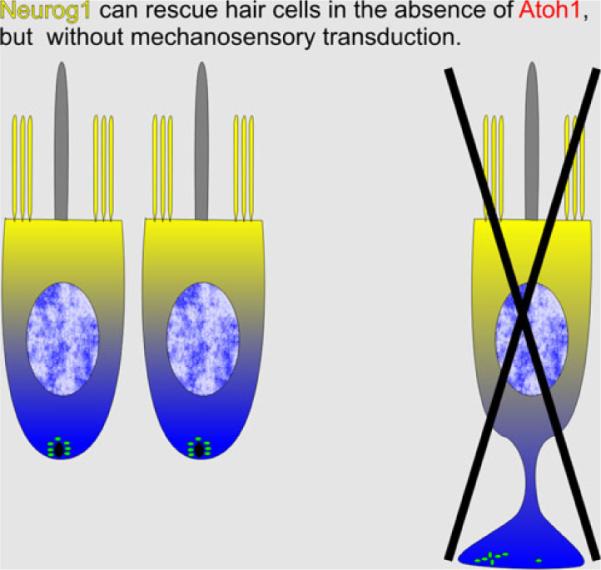
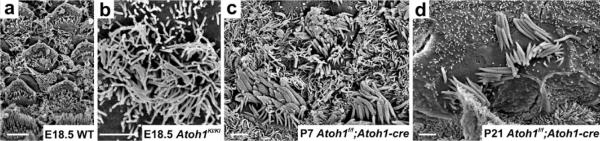
Expression of Neurog1 under Atoh1 promoter control results in partial differentiation of hair cell precursors. However, these cells rapidly degenerate without ever developing stereocilia. Instead, these cells develop central kinocilia surrounded by many microvilli. Despite a profound expression of Neurog1 and Neurod1 in these cells, none develop as neurons. This suggests that hair cell precursors are committed to differentiate as hair cells prior to Atoh1 upregulation and misexpression of a different bHLH gene cannot change that fate. This contrasts sharply with the fate change of sensory neurons in Neurod1 null mice shown in Fig. 5
Fig. 8.
Scanning electron microscopy images show abnormal OC cells with long microvilli surrounding a central kinocilium in the E18.5 mouse mutant where Atoh1 is replaced with Neurog1 (b). In the Atoh1 conditional knockout mouse mutant using Atoh1-cre to “self-terminate” after its limited transient expression, some apical hair cells develop stereocilia at an early postnatal stage (c). However, the stereocilia development is incomplete and transient, leading to further loss of stereocilia (d) and eventually loss of hair cells. This suggests that Atoh1 is necessary for both the initiation and the maintenance of the mechanotransduction development. Although the replacement of Atoh1 with Neurog1 can partially rescue the viability of the OC cells, Neurog1 can neither functionally support hair cell development nor transdifferentiate them into neurons. Bars (a) 10 μm, (b–d) 1 μm. The images are modified from Jahan et al. (2012), Pan et al. (2012)
In line with these differential effects of Atoh1 levels of expression in conjunction with co-expression of other bHLH genes, in particular Neurod1, are recent data on a conceptually novel mouse mutant, a “self-terminating” Atoh1 mouse (Pan et al. 2012). In this mouse, the Atoh1-cre transgene (Matei et al. 2005), which uses the previously isolated Atoh1 enhancer (Helms et al. 2000) to drive cre, is only activated after the floxed Atoh1 gene (Maricich et al. 2009) is transcribed into mRNA and translated into protein. The Atoh1-induced cre subsequently “self-terminates” continued transcription of Atoh1 mRNA by recombining the two floxed Atoh1 alleles. These mice show only a transient expression of Atoh1 mRNA that is never as profound as the expression in control littermates. Most interesting is the fate of the hair cell precursors. Despite only a transient expression of Atoh1, they initiate near normal differentiation, express Myo7a but fail to express Neurod1. The initial differentiation is followed by hair cell death with a variable time constant. Some hair cell precursors die within days whereas others remain for weeks. In particular, the IHCs die early whereas the first row of OHCs seems to have the longest viability. This effect indicates that continued but low level of Atoh1 expression (just enough to be detected using a LacZ reporter; Matei et al. 2005) is important for hair cell maintenance. Indeed, in Neurod1 mutant mice, Atoh1 expression remains elevated relative to control postnatal littermates, indicating that Neurod1 may function not only as a transcriptional activator but also as a repressor. If true, it indicates that overexpression of Atoh1 could ameliorate noise or chemically induced hair cell loss. A recent paper indicates that this might indeed be the case (He et al., in revision).
Inner versus outer hair cell identity may be defined by level and duration of Atoh1 expression
An optimized regenerative therapy that leads to the restoration of a functional OC will need the proper distribution and functional characteristics of IHCs and OHCs. As far as neurosensory subtype specification is concerned, many genes are differentially expressed in various subtypes of hair cells and supporting cells in the developed OC (Belyantseva et al. 2000; Montcouquiol and Kelley 2003; Zheng and Gao 1997). Consistent with these expression differences are differential defects in mutants (Deol 1981; Holley et al. 2010; Huh et al. 2012) or after chemical or noise ablations for IHCs and OHCs (Abrashkin et al. 2006). Best known is the differential distribution of Fgf8 in IHCs (Jacques et al. 2007; Pirvola et al. 2000) and Fgfr3 in pillar and Deiter's cells (Huh et al. 2012; Puligilla et al. 2007). Absence of Fgfr3 leads to alteration of pillar cell development including disorganization of type II afferent growth to OHCs (Puligilla et al. 2007), absence of Fgf20 results in loss of the outermost row of OHCs (Huh et al. 2012) and loss of Eya1/Six1 results in loss of all OHCs (Ahmed et al. 2012a). However, how Fgf8 is upregulated only in IHCs (Jahan et al. 2010b; Pirvola et al. 2000) and how Fgf20 and Eya1/Six1 manage to regulate formation of only some hair cells remain unclear. Despite the differential distribution of several markers in IHCs and OHCs (Cotanche and Kaiser 2010), no molecular steps are currently known that convert the general hair cell development, driven by Atoh1, toward differentiation into the two subtypes of hair cells found in all mammalian sensory epithelia. In fact, we do not yet understand at a molecular level how vestibular and OC hair cells are distinctly specified. However, studies in mutant mice suggest that segregation of sensory epithelia facilitates epithelia-specific hair cells differentially. This suggestion derives from data that show that lack of epithelia segregation results in overlapping distribution of vestibular and OC hair cells adjacent to each other (Nichols et al. 2008).
The only insight into this basic problem are recent data that suggest alterations in the interactions of bHLH TFs in developing neurosensory cells may play a role in cell subtype decision in the ear. Eliminating the bHLH gene Neurod1 results in transformation of OHCs into IHC-like cells that express Fgf8 and develop the thick stereocilia characteristic of IHCs (Jahan et al. 2010b). This transformation seems to come about through eliminating the Neurod1 mediated suppression of Atoh1 expression in the apex (Jahan et al. 2010b), suggesting that timing of Atoh1 expression, in combination with the level of its expression, plays a yet to be fully characterized role in the differentiation of IHCs and OHCs. Assessing this further requires the in vivo manipulation beyond the simple haploinsufficiency of Atoh1 (no phenotype has been reported for those mice) and the complete null of Atoh1. While Atoh1 null mice have no hair cells, much like after chemically induced ablation (Bermingham et al. 1999; Chen et al. 2002; Fritzsch et al. 2005), they are early lethal precluding any further study. Conditional mutant lines that eliminate Atoh1 prior to hair cell differentiation using, for example, Pax2-cre, can survive but have no OC remaining after birth and almost no sensory neurons (Pan et al. 2011). Our “self-terminating” Atoh1-cre:Atoh1 conditional deletion mice have a progressive and near complete loss of all IHCs shortly after birth but retain some OHCs in the apex until postnatal day 38. Interestingly, this mouse model retains many of the afferent and efferent innervations to the OC, mimicking a very severe early onset human presbycusis.
In these “self-terminating” mice, the IHCs are lost first (Pan et al. 2012). This is the opposite effect seen after Neurod1 loss, which mediates disinhibition of Atoh1 expression, which leads to transformation of OHCs into IHC-like cells (Jahan et al. 2010b). Combined, these two sets of data imply that it is not only the topology of Atoh1 expression that is essential for the initiation of hair cell differentiation in the sensory epithelia (and not in ganglia as in Neurod1 null mice) but also that both level and duration of expression play a major part in hair cell subtype specification in the ear. Our preliminary data obtained by combining the Neurog1 knockin allele (Jahan et al. 2012) with the self-terminating mouse model (Pan et al. 2012) directly test the hypothesis presented here. Consistent with our presumption, our data show an uncoupling of hair cell subtypes from their topology: cells in the position of inner or outer hair cells can differentiate either as inner or outer hair cells. Further analysis of these mice can provide the groundwork to align quantitative understanding of ear development with data in other systems where effects of intensity and duration of expression on cell fate decision are well known (Niwa et al. 2000; Pelet et al. 2011; Sansom et al. 2009), including quantitative effects of other bHLH genes (Conway et al. 2010).
In summary, studies in mouse ear development show a complex intra- and intercellular cross-regulation of the three atonal family bHLH TFs that indicates an incomplete segregation during development. We propose that this complex interplay reflects the evolutionary history of multiplying atonal family members and diversifying them to differentiate into two (hair cells and sensory neuron) instead of the single ancestral neurosensory cell. Combined, these three interacting atonal family members (and an undisclosed set of other bHLH TFs) are needed to generate the right number of the two neurosensory cell types. Intra- and intercellular cross-regulation between these TFs also affect the level and duration of expression of each factor and thereby determine not only cell type-specific differentiation but also help define the neurosensory subtypes. The formation of neurosensory cell types are largely a matter of temporal regulation (neurons first, followed later by hair cells in mice) possibly of clonally related cells (Fritzsch et al. 2006), while the subtype specification depends on both the time and intensity of expression of these TFs, one of the least understood aspects of gene expression regulation (Crocker et al. 2008).
Remarkably, 600 million years after the vertebrate ancestors evolved the interactive cascade of three atonal family members to guide the development of two cell types in the ear and the lateral line system, there is still overlapping expression and cross-regulation within cells suggestive of incomplete segregation of their promoter-mediated expression. Both spatial and temporal expression changes enhance the degree of segregation of sensory neuron precursors from hair cell precursors but do not eliminate them. Whether this remaining overlap is under positive selection or simply reflects a historic anachronism that will eventually disappear over evolutionary time remains to be explored.
What regulates Atoh1, Neurog1 and Neurod1 expression in the ear and how much does their downstream gene repertoire overlap?
From the above insight into the evolutionary and developmental dynamics of inner ear bHLH gene expression and their effects on various neurosensory cell types in the ear, it should be clear that the regulation of these TFs has to be tightly controlled in space, time and intensity. While the above-outlined data, combined with previous analysis of promoter regions of these genes (Ahmed et al. 2012a, b; Gowan et al. 2001; Helms et al. 2000; Quinones et al. 2010), suggest an intricate intracellular and extracellular cross-regulation, it is unclear what is regulating the highly patterned initial expression to specific cells of the developing prosensory epithelia but diffusible factors such as Fgfs and Bmps are likely candidates (Fritzsch et al. 2006). It is clear that Neurog1 is the first bHLH gene to be expressed at embryonic day (E) 8.75 in the mouse (Ma et al. 1998) followed by Neurod1, which is evidently regulated by Neurog1 (Jahan et al. 2012; Ma et al. 1998). The last of these TFs to be expressed is Atoh1, which makes its appearance at E10.5 as revealed by RT-qPCR (Matei et al. 2005). Apparently, expression of Neurod1 in hair cells is even further delayed as it is dependent on Atoh1 for its expression, while it inhibits Atoh1 in a negative feedback loop of unknown complexity (Jahan et al. 2010b; Matei et al. 2005).
Several candidate genes can be proposed as upstream regulators of these three atonal family members based on data generated in insects (Garcia-Bellido and de Celis 2009) but none of them have been tested in vertebrates and thus remain speculative at the moment. Multiple TFs have been identified in ear development that are needed for hair cell and neuronal development (Ahmed et al. 2012a, b; Bouchard et al. 2010; Duncan et al. 2011) but their interaction remains to be elucidated. The various TFs needed for neurosensory cell precursors to respond to expression of a given bHLH TF with proper differentiation should not be confused with those factors regulating the expression of bHLH TFs. Clearly, the Delta/Notch system is an essential stabilizer of the differentiation pattern of the OC (Adam et al. 1998; Brooker et al. 2006; Daudet et al. 2007; Pan et al. 2010; Zine et al. 2001). However, the entire Delta/Notch signaling cascade can be blocked without affecting initial proneural gene upregulation in specific positions (Basch et al. 2011). Indeed, the first publication describing the effect of eliminating a proneural gene in the ear already showed that the Delta/Notch expression is dependent on the expression of Neurog1 and is not inducing it (Ma et al. 1998). Without knowing exactly which TFs combine to regulate topographically restricted proneural gene expression in the ear, we will not be able to design ways to regulate their expression in vivo through selective activation of their promoter regions to restore an OC. Some data suggest that multiple TFs with a complex interaction in Atoh1 promoter regions are needed for that expression (Ahmed et al. 2012a), without detailing the additional TFs already characterized in the Atoh1 promoter region required to drive Atoh1 in a topologically restricted fashion such as Gata3 (Duncan et al. 2011) or Pax2 (Bouchard et al. 2010).
The known aspects of promoter regions of these three bHLH genes are composed of multiple TF binding sites, including E-boxes for self-binding and binding of other related bHLH TFs (Ahmed et al. 2012a). Atoh1 binds to an E-box consensus binding site in its enhancer and autor-egulates its expression (Helms et al. 2000). In addition, the Atoh1 enhancer also contains a second E-box that is highly conserved. But the binding factor has not been identified and its role in regulating Atoh1 expression is unknown. The promoters of Neurod1 and Neurog1 have not been fully characterized; however, it has been suggested that the activity of remote regulatory sequences may direct the spatio-temporally specific expression of these genes (Gowan et al. 2001). Further studies to identify and characterize these regulatory elements and the binding factors will also provide the molecular mechanism for the intracellular cross-regulation of these bHLH genes.
In the absence of identified genes that could directly mediate the bHLH gene activation in vivo through binding to their promoters, the only alternative for promoter regulation-driven gene activation would be the direct activation of relevant downstream genes. However, studies in the nervous system have shown that each of the three relevant bHLH TFs has several hundred downstream target genes (Dalgard et al. 2011; Klisch et al. 2011; Seo et al. 2007). Roughly half of them are present in the ear during different development stages (Sajan et al. 2007, 2011) (Fig. 9). Studies in the ear may identify even more ear-specific targets for these bHLH genes. Which ones are the most important to elicit a cascading effect remains unclear. Obviously, understanding the interaction of TFs to regulate the space, intensity and duration of Atoh1 expression might be the more logical way to go, despite its emerging complexity (Ahmed et al. 2012a).
Fig. 9.
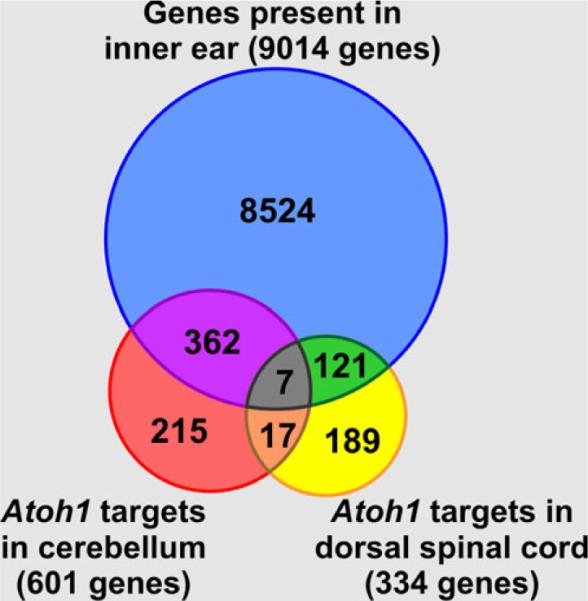
Atoh1 downstream targets identified in the cerebellum and the dorsal spinal cord were compared with the list of genes that are expressed in the inner ear; 61 % of the Atoh1 target genes in the cerebellum and 38 % in the dorsal spinal cord are also expressed and potentially regulated by Atoh1 in the inner ear
How can these insights translate into treatment to help against hearing loss?
There are still several hurdles to overcome for an effective translation of these molecular insights into treatment. If we take the approach that the main problem we need to understand is the topologically restricted upregulation of a few TFs that induce hair cells directly through transdifferentiation (Yoo et al. 2011), or allow transfection with Atoh1 to effectively differentiate new hair cells, or allow hair cells generated in vitro to differentiate appropriately, we can then begin to work on genetically engineered mouse models that allow us to test certain premises. Alternatively, we can use the molecular knowledge at hand and try, for example, to transform the basilar papilla of frogs (Fritzsch and Wake 1988) into a mammalian type of organ using modern genetic techniques now possible in these animals (Abu-Daya et al. 2012), thus demonstrating that we indeed understand all the relevant genes.
When confronted with a complete loss of an OC and replacement by a flat epithelium (Izumikawa et al. 2008; Pan et al. 2011), there is no clear path to success in the foreseeable future. However, such cases of complete loss of all hair cells are mostly limited to treatment with ototoxic drugs in humans, as in experimental animals. In fact, most hearing loss, in particular age-related hearing loss, does not lead to a sudden and near complete loss of all hair cells. While several mouse lines show signs of presbycusis, the molecular basis of these effects is typically as unknown as the genetic predisposition is for presbycusis in humans. Like in these mouse lines, the common phenotype of hair cell loss in humans with presbycusis is probably the convergence of a multitude of minor genetic defects that require several genome-wide association studies to narrow them down. The unclear etiology notwithstanding, generating mouse lines that are genetically modified to show progressive hair cell loss with specific loss of one subtype of hair cells could help provide mouse models for human presbycusis. Several mouse models (see below) could allow insights toward curing the progressive hearing loss through restoration of the lost hair cells while maintaining the remaining hair cells. Such data could provide a beginning to reveal manipulations to ensure proper hair cell subtype development in cases with a more complete loss of hair cells.
Furthermore, in contrast to the past animal models with a chemically induced complete loss of all hair cells, genetically engineered mouse models could demonstrate whether the remaining hair cells can function as a template to help organize the differentiation of hair cells into two subtypes in the right position and thus restore hearing. Such a mouse model should, therefore, have only a partial loss, preferentially only of one subtype of hair cells, with retention of others that may or may not be differentiated fully as hair cells. Likewise, these mice should also show a progressive and complete loss of all hair cells over time but should retain most of the sensory neurons to assess the function of a restored OC. The complete loss over time is essential to verify the effectiveness of reconstitution of hair cells and their retention. Existing mouse models with a knockout of genes relevant for hair cell maintenance such as Pou4f3 (previously Brn3c) or Barhl1 (Chellappa et al. 2008; Xiang et al. 2003) are too complex to be useful here, as they require not only the restoration of hair cells but in addition the full functional replacement of the knockout genes, as otherwise the regenerated hair cells will degenerate again. Essentially, this model would need to have a progressive hair cell loss driven by the loss of the same gene that is typically used to restore hair cells, Atoh1.
Unfortunately, Atoh1 null mice are early lethal and have also a complete loss of all hair cells much like chemically induced ablations (Bermingham et al. 1999; Chen et al. 2002; Fritzsch et al. 2005). Conditional mutant lines that eliminate Atoh1 prior to hair cell differentiation using, for example, Pax2-cre, can survive but have no OC left and almost no sensory neurons (Pan et al. 2011). Using an available Atoh1-cre line (Matei et al. 2005), we genetically engineered a “self-terminating” Atoh1 expression (Pan et al. 2012). This mouse model has a progressive and near complete loss of all IHCs shortly after birth, retains only a few OHCs in the apex at postnatal day 38 but retains most of the afferent and efferent innervation (Pan et al. 2012). Most importantly, these mice lose all endogenous Atoh1 in remaining hair cells due to the self-termination mechanism whereby the Atoh1 protein activates the Atoh1 enhancer-mediated cre expression that recombines the floxed Atoh1 gene (Pan et al. 2012).
These new mice are an ideal genetically engineered model for attempts to induce novel hair cell differentiation, rescuing otherwise slowly dying hair cells, using established protocols for adenoviral transfection (Izumikawa et al. 2005, 2008; Staecker et al. 2011). Using adenovirus-mediated Atoh1 transfection, one can directly test in these mice if the remaining undifferentiated Myo7a-positive “hair cells” initiate differentiation into functional hair cells that remain viable over a longer period. This mouse model can also help to understand how duration and intensity of a virally mediated Atoh1 expression relates to hair cell subtype differentiation independent of their position. Therefore, combining the genetically engineered mouse with the adenoviral transfection approach, one could critically test the hypothesis that the level and duration of Atoh1 determine not only overall viability but also the subtype of hair cells. Such a test would also validate whether existing hair cells can serve as a template to expand the residual OC to fill in hair cells between patches of remaining hair cells or to fill in gaps in remaining rows of existing hair cells. If such effects occur, it could lead to a “replenishment therapy” prior to the complete loss of the OC, an approach that could benefit particularly patients with slow progressing age-related hearing loss.
In summary, understanding how non-mammals can replace hair cells at a molecular level does not by itself guarantee transferability to induce similar processes in humans. In contrast, recently molecularly engineered mice are an ideal model for attempts to induce novel hair cell differentiation of slowly dying hair cells using established protocols for adenoviral transfection (Izumikawa et al. 2005, 2008) with a high level of translatability to humans. Using these mice (Pan et al. 2012), one could establish how the transient expression of a limited amount of Atoh1 translates at a molecular level to a variable hair cell loss over time. This will also test whether existing hair cells can serve as a template to expand the residual OC to fill in gaps between patches of remaining hair cells.
Acknowledgments
This work was supported by NIH grants R01 DC 005590 (to B.F.), CTSA UL1RR024979 (to B.K.) and P30 DC 010362 (http://www.nidcd.nih.gov). We also acknowledge the support from the Office of the Vice President for Research (https://research.uiowa.edu/ovpr/office-vp-research) and College of Liberal Arts & Sciences (http://www.clas.uiowa.edu) at the University of Iowa.
References
- Abrashkin KA, Izumikawa M, Miyazawa T, Wang CH, Crumling MA, Swiderski DL, Beyer LA, Gong TW, Raphael Y. The fate of outer hair cells after acoustic or ototoxic insults. Hear Res. 2006;218:20–29. doi: 10.1016/j.heares.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Abu-Daya A, Khokha MK, Zimmerman LB. The hitchhiker's guide to Xenopus genetics. Genesis. 2012;50:164–175. doi: 10.1002/dvg.22007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam J, Myat A, Le Roux I, Eddison M, Henrique D, Ish-Horowicz D, Lewis J. Cell fate choices and the expression of Notch, Delta and Serrate homologues in the chick inner ear: parallels with Drosophila sense-organ development. Development. 1998;125:4645–4654. doi: 10.1242/dev.125.23.4645. [DOI] [PubMed] [Google Scholar]
- Ahmed M, Wong EYM, Sun J, Xu J, Wang F, Xu P-X. Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev Cell. 2012a;22:377–390. doi: 10.1016/j.devcel.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed M, Xu J, Xu PX. EYA1 and SIX1 drive the neuronal developmental program in cooperation with the SWI/SNF chromatin-remodeling complex and SOX2 in the mammalian inner ear. Development. 2012b doi: 10.1242/dev.071670. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SA, Robinson BK, Huang J, Green SH. Prosurvival and proapoptotic intracellular signaling in rat spiral ganglion neurons in vivo after the loss of hair cells. J Comp Neurol. 2007;503:832–852. doi: 10.1002/cne.21430. [DOI] [PubMed] [Google Scholar]
- Ali F, Hindley C, McDowell G, Deibler R, Jones A, Kirschner M, Guillemot F, Philpott A. Cell cycle-regulated multi-site phosphorylation of Neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development. 2011;138:4267–4277. doi: 10.1242/dev.067900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basch ML, Ohyama T, Segil N, Groves AK. Canonical Notch signaling is not necessary for prosensory induction in the mouse cochlea: insights from a conditional mutant of RBPjkappa. J Neurosci. 2011;31:8046–8058. doi: 10.1523/JNEUROSCI.6671-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batts SA, Shoemaker CR, Raphael Y. Notch signaling and Hes labeling in the normal and drug-damaged organ of Corti. Hear Res. 2009;249:15–22. doi: 10.1016/j.heares.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyantseva IA, Adler HJ, Curi R, Frolenkov GI, Kachar B. Expression and localization of prestin and the sugar transporter GLUT-5 during development of electromotility in cochlear outer hair cells. J Neurosci. 2000;20:RC116. doi: 10.1523/JNEUROSCI.20-24-j0002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- Bok J, Dolson DK, Hill P, Ruther U, Epstein DJ, Wu DK. Opposing gradients of Gli repressor and activators mediate Shh signaling along the dorsoventral axis of the inner ear. Development. 2007;134:1713–1722. doi: 10.1242/dev.000760. [DOI] [PubMed] [Google Scholar]
- Bouchard M, de Caprona D, Busslinger M, Xu P, Fritzsch B. Pax2 and Pax8 cooperate in mouse inner ear morphogenesis and innervation. BMC Dev Biol. 2010;10:89. doi: 10.1186/1471-213X-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2011 doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgham JT, Carroll SM, Thornton JW. Evolution of hormone-receptor complexity by molecular exploitation. Science. 2006;312:97–101. doi: 10.1126/science.1123348. [DOI] [PubMed] [Google Scholar]
- Brigande JV, Heller S. Quo vadis, hair cell regeneration? Nat Neurosci. 2009;12:679–685. doi: 10.1038/nn.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006;133:1277–1286. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
- Budelmann B. Hearing in nonarthropod invertebrates. In: Webster DB, Fay RR, Popper AN, editors. The evolutionary biology of hearing. Springer; New York: 1992. pp. 141–155. [Google Scholar]
- Burighel P, Caicci F, Manni L. Hair cells in non-vertebrate models: lower chordates and molluscs. Hear Res. 2011;273:14–24. doi: 10.1016/j.heares.2010.03.087. [DOI] [PubMed] [Google Scholar]
- Cachero S, Simpson TI, Zur Lage PI, Ma L, Newton FG, Holohan EE, Armstrong JD, Jarman AP. The gene regulatory cascade linking proneural specification with differentiation in Drosophila sensory neurons. PLoS Biol. 2011;9:e1000568. doi: 10.1371/journal.pbio.1000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Lin Z, Kulessa H, Hebert J, Hogan BL, Wu DK. Bmp4 is essential for the formation of the vestibular apparatus that detects angular head movements. PLoS Genet. 2008;4:e1000050. doi: 10.1371/journal.pgen.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappa R, Li S, Pauley S, Jahan I, Jin K, Xiang M. Barhl1 regulatory sequences required for cell-specific gene expression and autoregulation in the inner ear and central nervous system. Mol Cell Biol. 2008;28:1905–1914. doi: 10.1128/MCB.01454-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Johnson JE, Zoghbi HY, Segil N. The role of Math1 in inner ear development: Uncoupling the establishment of the sensory primordium from hair cell fate determination. Development. 2002;129:2495–2505. doi: 10.1242/dev.129.10.2495. [DOI] [PubMed] [Google Scholar]
- Conway SJ, Firulli B, Firulli AB. A bHLH code for cardiac morphogenesis. Pediatr Cardiol. 2010;31:318–324. doi: 10.1007/s00246-009-9608-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotanche DA, Kaiser CL. Hair cell fate decisions in cochlear development and regeneration. Hear Res. 2010;266:18–25. doi: 10.1016/j.heares.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker J, Tamori Y, Erives A. Evolution acts on enhancer organization to fine-tune gradient threshold readouts. PLoS Biol. 2008;6:e263. doi: 10.1371/journal.pbio.0060263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabdoub A, Puligilla C, Jones JM, Fritzsch B, Cheah KS, Pevny LH, Kelley MW. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci USA. 2008;105:18396–18401. doi: 10.1073/pnas.0808175105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgard CL, Zhou Q, Lundell TG, Doughty ML. Altered gene expression in the emerging cerebellar primordium of Neurog1−/−mice. Brain Res. 2011;1388:12–21. doi: 10.1016/j.brainres.2011.02.087. [DOI] [PubMed] [Google Scholar]
- Daudet N, Ariza-McNaughton L, Lewis J. Notch signalling is needed to maintain, but not to initiate, the formation of prosensory patches in the chick inner ear. Development. 2007;134:2369–2378. doi: 10.1242/dev.001842. [DOI] [PubMed] [Google Scholar]
- Degnan BM, Vervoort M, Larroux C, Richards GS. Early evolution of metazoan transcription factors. Curr Opin Genet Dev. 2009;19:591–599. doi: 10.1016/j.gde.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Deol MS. The inner ear in Bronx waltzer mice. Acta Otolaryngol. 1981;92:331–336. doi: 10.3109/00016488109133269. [DOI] [PubMed] [Google Scholar]
- Doetzlhofer A, Basch ML, Ohyama T, Gessler M, Groves AK, Segil N. Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev Cell. 2009;16:58–69. doi: 10.1016/j.devcel.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Frutos E, Lopez-Hernandez I, Vendrell V, Neves J, Gallozzi M, Gutsche K, Quintana L, Sharpe J, Knoepfler PS, Eisenman RN, Trumpp A, Giraldez F, Schimmang T. N-myc controls proliferation, morphogenesis, and patterning of the inner ear. J Neurosci. 2011;31:7178–7189. doi: 10.1523/JNEUROSCI.0785-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JS, Lim KC, Engel JD, Fritzsch B. Limited inner ear morphogenesis and neurosensory development are possible in the absence of GATA3. Int J Dev Biol. 2011;55:297–303. doi: 10.1387/ijdb.103178jd. [DOI] [PubMed] [Google Scholar]
- Espinosa-Soto C, Martin OC, Wagner A. Phenotypic plasticity can facilitate adaptive evolution in gene regulatory circuits. BMC Evol Biol. 2011;11:5. doi: 10.1186/1471-2148-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Wake MH. The inner ear of gymnophione amphibians and its nerve supply: a comparative study of regressive events in a complex sensory system. Zoomorphology. 1988;108:210–217. [Google Scholar]
- Fritzsch B, Beisel KW, Bermingham NA. Developmental evolutionary biology of the vertebrate ear: conserving mechanoelectric transduction and developmental pathways in diverging morphologies. Neuroreport. 2000;11:R35–44. doi: 10.1097/00001756-200011270-00013. [DOI] [PubMed] [Google Scholar]
- Fritzsch B, Beisel KW, Jones K, Farinas I, Maklad A, Lee J, Reichardt LF. Development and evolution of inner ear sensory epithelia and their innervation. J Neurobiol. 2002;53:143–156. doi: 10.1002/neu.10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Matei VA, Nichols DH, Bermingham N, Jones K, Beisel KW, Wang VY. Atoh1 null mice show directed afferent fiber growth to undifferentiated ear sensory epithelia followed by incomplete fiber retention. Dev Dyn. 2005;233:570–583. doi: 10.1002/dvdy.20370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Beisel KW, Hansen LA. The molecular basis of neurosensory cell formation in ear development: a blueprint for hair cell and sensory neuron regeneration? Bioessays. 2006;28:1181–1193. doi: 10.1002/bies.20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Beisel KW, Pauley S, Soukup G. Molecular evolution of the vertebrate mechanosensory cell and ear. Int J Dev Biol. 2007;51:663–678. doi: 10.1387/ijdb.072367bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Eberl DF, Beisel KW. The role of bHLH genes in ear development and evolution: revisiting a 10-year-old hypothesis. Cell Mol Life Sci. 2010;67:3089–3099. doi: 10.1007/s00018-010-0403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Jahan I, Pan N, Kersigo J, Duncan J, Kopecky B. Dissecting the molecular basis of organ of Corti development: Where are we now? Hear Res. 2011;276:16–26. doi: 10.1016/j.heares.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliot B, Quiquand M. A two-step process in the emergence of neurogenesis. Eur J Neurosci. 2011;34:847–862. doi: 10.1111/j.1460-9568.2011.07829.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido A, de Celis JF. The complex tale of the achaetescute complex: a paradigmatic case in the analysis of gene organization and function during development. Genetics. 2009;182:631–639. doi: 10.1534/genetics.109.104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazave E, Lapebie P, Richards GS, Brunet F, Ereskovsky AV, Degnan BM, Borchiellini C, Vervoort M, Renard E. Origin and evolution of the Notch signalling pathway: an overview from eukaryotic genomes. BMC Evol Biol. 2009;9:249. doi: 10.1186/1471-2148-9-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowan K, Helms AW, Hunsaker TL, Collisson T, Ebert PJ, Odom R, Johnson JE. Crossinhibitory activities of Ngn1 and Math1 allow specification of distinct dorsal interneurons. Neuron. 2001;31:219–232. doi: 10.1016/s0896-6273(01)00367-1. [DOI] [PubMed] [Google Scholar]
- Groves AK. The challenge of hair cell regeneration. Exp Biol Med (Maywood) 2010;235:434–446. doi: 10.1258/ebm.2009.009281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves AK, Fekete DM. Shaping sound in space: the regulation of inner ear patterning. Development. 2012;139:245–257. doi: 10.1242/dev.067074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- Holley M, Rhodes C, Kneebone A, Herde MK, Fleming M, Steel KP. Emx2 and early hair cell development in the mouse inner ear. Dev Biol. 2010;340:547–556. doi: 10.1016/j.ydbio.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Sage C, Tang Y, Lee SG, Petrillo M, Hinds PW, Chen ZY. Overlapping and distinct pRb pathways in the mammalian auditory and vestibular organs. Cell Cycle. 2011;10:337–351. doi: 10.4161/cc.10.2.14640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh SH, Jones J, Warchol ME, Ornitz DM. Differentiation of the lateral compartment of the cochlea requires a temporally restricted FGF20 signal. PLoS Biol. 2012;10:e1001231. doi: 10.1371/journal.pbio.1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumikawa M, Minoda R, Kawamoto K, Abrashkin KA, Swiderski DL, Dolan DF, Brough DE, Raphael Y. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat Med. 2005;11:271–276. doi: 10.1038/nm1193. [DOI] [PubMed] [Google Scholar]
- Izumikawa M, Batts SA, Miyazawa T, Swiderski DL, Raphael Y. Response of the flat cochlear epithelium to forced expression of Atoh1. Hear Res. 2008;240:52–56. doi: 10.1016/j.heares.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques BE, Montcouquiol ME, Layman EM, Lewandoski M, Kelley MW. Fgf8 induces pillar cell fate and regulates cellular patterning in the mammalian cochlea. Development. 2007;134:3021–3029. doi: 10.1242/dev.02874. [DOI] [PubMed] [Google Scholar]
- Jahan I, Kersigo J, Pan N, Fritzsch B. Neurod1 regulates survival and formation of connections in mouse ear and brain. Cell Tissue Res. 2010a;341:95–110. doi: 10.1007/s00441-010-0984-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan I, Pan N, Kersigo J, Fritzsch B. Neurod1 suppresses hair cell differentiation in ear ganglia and regulates hair cell subtype development in the cochlea. PLoS One. 2010b;5:e11661. doi: 10.1371/journal.pone.0011661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan I, Pan N, Kersigo J, Calisto LE, Morris KA, Kopecky B, Duncan JS, Beiseld KW, Fritzsch B. Expression of Neurog1 instead of Atoh1 can partially rescue organ of Corti cell survival. PLoS One. 2012;7:e30853. doi: 10.1371/journal.pone.0030853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon SJ, Fujioka M, Kim SC, Edge AS. Notch signaling alters sensory or neuronal cell fate specification of inner ear stem cells. J Neurosci. 2011;31:8351–8358. doi: 10.1523/JNEUROSCI.6366-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen JM. Evolution of octavolateralis sensory cells. In: Coombs S, Goerner P, Muenz H, editors. The mechanosensory lateral line. Neurobiology and evolution. Springer; New York: 1989. pp. 99–115. [Google Scholar]
- Jungebluth P, Macchiarini P. Stem cell-based therapy and regenerative approaches to diseases of the respiratory system. Br Med Bull. 2011;99:169–187. doi: 10.1093/bmb/ldr028. [DOI] [PubMed] [Google Scholar]
- Jungebluth P, Alici E, Baiguera S, Le Blanc K, Blomberg P, Bozoky B, Crowley C, Einarsson O, Grinnemo KH, Gudbjartsson T, Le Guyader S, Henriksson G, Hermanson O, Juto JE, Leidner B, Lilja T, Liska J, Luedde T, Lundin V, Moll G, Nilsson B, Roderburg C, Stromblad S, Sutlu T, Teixeira AI, Watz E, Seifalian A, Macchiarini P. Tracheobronchial transplantation with a stem-cell-seeded bioartificial nanocomposite: a proof-of-concept study. Lancet. 2011;378:1997–2004. doi: 10.1016/S0140-6736(11)61715-7. [DOI] [PubMed] [Google Scholar]
- Karis A, Pata I, van Doorninck JH, Grosveld F, de Zeeuw CI, de Caprona D, Fritzsch B. Transcription factor GATA-3 alters pathway selection of olivocochlear neurons and affects morpho-genesis of the ear. J Comp Neurol. 2001;429:615–630. doi: 10.1002/1096-9861(20010122)429:4<615::aid-cne8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Kiernan AE, Xu J, Gridley T. The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet. 2006;2:e4. doi: 10.1371/journal.pgen.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Fritzsch B, Serls A, Bakel LA, Huang EJ, Reichardt LF, Barth DS, Lee JE. NeuroD-null mice are deaf due to a severe loss of the inner ear sensory neurons during development. Development. 2001;128:417–426. doi: 10.1242/dev.128.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klisch TJ, Xi Y, Flora A, Wang L, Li W, Zoghbi HY. In vivo Atoh1 targetome reveals how a proneural transcription factor regulates cerebellar development. Proc Natl Acad Sci USA. 2011;108:3288–3293. doi: 10.1073/pnas.1100230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky B, Fritzsch B. Regeneration of hair cells: making sense of all the noise. Pharm (Basel) 2011;4:848–879. doi: 10.3390/ph4060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky B, Santi P, Johnson S, Schmitz H, Fritzsch B. Conditional deletion of N-Myc disrupts neurosensory and non-sensory development of the ear. Dev Dyn. 2011;240:1373–1390. doi: 10.1002/dvdy.22620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmik Z, Daube M, Frei E, Norman B, Kos L, Dishaw LJ, Noll M, Piatigorsky J. Role of pax genes in eye evolution. A Cnidarian PaxB gene uniting Pax2 and Pax6 functions. Dev Cell. 2003;5:773–785. doi: 10.1016/s1534-5807(03)00325-3. [DOI] [PubMed] [Google Scholar]
- Kruger M, Schmid T, Kruger S, Bober E, Braun T. Functional redundancy of NSCL-1 and NeuroD during development of the petrosal and vestibulocochlear ganglia. Eur J Neurosci. 2006;24:1581–1590. doi: 10.1111/j.1460-9568.2006.05051.x. [DOI] [PubMed] [Google Scholar]
- Laine H, Doetzlhofer A, Mantela J, Ylikoski J, Laiho M, Roussel MF, Segil N, Pirvola U. p19(Ink4d) and p21(Cip1) collaborate to maintain the postmitotic state of auditory hair cells, their codeletion leading to DNA damage and p53-mediated apoptosis. J Neurosci. 2007;27:1434–1444. doi: 10.1523/JNEUROSCI.4956-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander AD. Pattern, growth, and control. Cell. 2011;144:955–969. doi: 10.1016/j.cell.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MP, Yutzey KE. Twist1 directly regulates genes that promote cell proliferation and migration in developing heart valves. PLoS One. 2011;6:e29758. doi: 10.1371/journal.pone.0029758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HJ, Ray SK, Singh NK, Johnston B, Leiter AB. Basic helix-loop-helix transcription factors and enteroendocrine cell differentiation. Diabetes Obes Metab. 2011;13(Suppl 1):5–12. doi: 10.1111/j.1463-1326.2011.01438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zuo J. Cell cycle regulation in hair cell development and regeneration in the mouse cochlea. Cell Cycle. 2008;7:2129–2133. doi: 10.4161/cc.7.14.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loponen H, Ylikoski J, Albrecht JH, Pirvola U. Restrictions in cell cycle progression of adult vestibular supporting cells in response to ectopic cyclin D1 expression. PLoS One. 2011;6:e27360. doi: 10.1371/journal.pone.0027360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan E, Chanda S, Ahlenius H, Sudhof TC, Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc Natl Acad Sci USA. 2012 doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Chen Z, del Barco BI, de la Pompa JL, Anderson DJ. neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;20:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- Ma Q, Anderson DJ, Fritzsch B. Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. J Assoc Res Otolaryngol. 2000;1:129–143. doi: 10.1007/s101620010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantela J, Jiang Z, Ylikoski J, Fritzsch B, Zacksenhaus E, Pirvola U. The retinoblastoma gene pathway regulates the postmitotic state of hair cells of the mouse inner ear. Development. 2005;132:2377–2388. doi: 10.1242/dev.01834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricich SM, Xia A, Mathes EL, Wang VY, Oghalai JS, Fritzsch B, Zoghbi HY. Atoh1-lineal neurons are required for hearing and for the survival of neurons in the spiral ganglion and brain-stem accessory auditory nuclei. J Neurosci. 2009;29:11123–11133. doi: 10.1523/JNEUROSCI.2232-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matei V, Pauley S, Kaing S, Rowitch D, Beisel KW, Morris K, Feng F, Jones K, Lee J, Fritzsch B. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev Dyn. 2005;234:633–650. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoda R, Izumikawa M, Kawamoto K, Zhang H, Raphael Y. Manipulating cell cycle regulation in the mature cochlea. Hear Res. 2007;232:44–51. doi: 10.1016/j.heares.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montcouquiol M, Kelley MW. Planar and vertical signals control cellular differentiation and patterning in the mammalian cochlea. J Neurosci. 2003;23:9469–9478. doi: 10.1523/JNEUROSCI.23-28-09469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols DH, Pauley S, Jahan I, Beisel KW, Millen KJ, Fritzsch B. Lmx1a is required for segregation of sensory epithelia and normal ear histogenesis and morphogenesis. Cell Tissue Res. 2008;334:339–358. doi: 10.1007/s00441-008-0709-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- O'Brien EK, Degnan B. Expression of Pax258 in the gastropod statocyst: insights into the antiquity of metazoan geosensory organs. Evol Dev. 2003;5:572–578. doi: 10.1046/j.1525-142x.2003.03062.x. [DOI] [PubMed] [Google Scholar]
- Oesterle EC, Chien WM, Campbell S, Nellimarla P, Fero ML. p27(Kip1) is required to maintain proliferative quiescence in the adult cochlea and pituitary. Cell Cycle. 2011;10:1237–1248. doi: 10.4161/cc.10.8.15301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama T, Groves AK. Expression of mouse Foxi class genes in early craniofacial development. Dev Dyn. 2004;231:640–646. doi: 10.1002/dvdy.20160. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Basch ML, Mishina Y, Lyons KM, Segil N, Groves AK. BMP signaling is necessary for patterning the sensory and nonsensory regions of the developing mammalian cochlea. J Neurosci. 2011;30:15044–15051. doi: 10.1523/JNEUROSCI.3547-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Nakagawa T, Kojima K, Matsumoto M, Kawauchi T, Hoshino M, Ito J. Silencing p27 reverses post-mitotic state of supporting cells in neonatal mouse cochleae. Mol Cell Neurosci. 2009;42:391–398. doi: 10.1016/j.mcn.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Oshima K, Shin K, Diensthuber M, Peng AW, Ricci AJ, Heller S. Mechanosensitive hair cell-like cells from embryonic and induced pluripotent stem cells. Cell. 2010;141:704–716. doi: 10.1016/j.cell.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeki M, Hamajima Y, Feng L, Ondrey FG, Schlentz E, Lin J. Id1 induces the proliferation of cochlear sensory epithelial cells via the nuclear factor-kappaB/cyclin D1 pathway in vitro. J Neurosci Res. 2007;85:515–524. doi: 10.1002/jnr.21133. [DOI] [PubMed] [Google Scholar]
- Pan W, Jin Y, Stanger B, Kiernan AE. Notch signaling is required for the generation of hair cells and supporting cells in the mammalian inner ear. Proc Natl Acad Sci USA. 2010;107:15798–15803. doi: 10.1073/pnas.1003089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan N, Jahan I, Kersigo J, Kopecky B, Santi P, Johnson S, Schmitz H, Fritzsch B. Conditional deletion of Atoh1 using Pax2-Cre results in viable mice without differentiated cochlear hair cells that have lost most of the organ of Corti. Hear Res. 2011;275:66–80. doi: 10.1016/j.heares.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan N, Jahan I, Kersigo J, Duncan J, Kopecky B, Fritzsch B. A novel Atoh1 'self-terminating' mouse model reveals the necessity of proper Atoh1 expression level and duration for inner ear hair cell differentiation and viability. PLoS One. 2012;7:e30358. doi: 10.1371/journal.pone.0030358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, Wernig M. Induction of human neuronal cells by defined transcription factors. Nature advance online publication. 2011 doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley S, Wright TJ, Pirvola U, Ornitz D, Beisel K, Fritzsch B. Expression and function of FGF10 in mammalian inner ear development. Dev Dyn. 2003;227:203–215. doi: 10.1002/dvdy.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley S, Lai E, Fritzsch B. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev Dyn. 2006;235:2470–2482. doi: 10.1002/dvdy.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelet S, Rudolf F, Nadal-Ribelles M, de Nadal E, Posas F, Peter M. Transient activation of the HOG MAPK pathway regulates bimodal gene expression. Science. 2011;332:732–735. doi: 10.1126/science.1198851. [DOI] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. Evolution of gene regulatory networks controlling body plan development. Cell. 2011;144:970–985. doi: 10.1016/j.cell.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirvola U, Spencer-Dene B, Xing-Qun L, Kettunen P, Thesleff I, Fritzsch B, Dickson C, Ylikoski J. FGF/FGFR-2(IIIb) signaling is essential for inner ear morphogenesis. J Neurosci. 2000;20:6125–6134. doi: 10.1523/JNEUROSCI.20-16-06125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puligilla C, Kelley MW. Building the world's best hearing aid; regulation of cell fate in the cochlea. Curr Opin Genet Dev. 2009;19:368–373. doi: 10.1016/j.gde.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puligilla C, Feng F, Ishikawa K, Bertuzzi S, Dabdoub A, Griffith AJ, Fritzsch B, Kelley MW. Disruption of fibroblast growth factor receptor 3 signaling results in defects in cellular differentiation, neuronal patterning, and hearing impairment. Dev Dyn. 2007;236:1905–1917. doi: 10.1002/dvdy.21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones HI, Savage TK, Battiste J, Johnson JE. Neurogenin 1 (Neurog1) expression in the ventral neural tube is mediated by a distinct enhancer and preferentially marks ventral interneuron lineages. Dev Biol. 2010;340:283–292. doi: 10.1016/j.ydbio.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raft S, Koundakjian EJ, Quinones H, Jayasena CS, Goodrich LV, Johnson JE, Segil N, Groves AK. Cross-regulation of Ngn1 and Math1 coordinates the production of neurons and sensory hair cells during inner ear development. Development. 2007;134:4405–4415. doi: 10.1242/dev.009118. [DOI] [PubMed] [Google Scholar]
- Riccomagno MM, Martinu L, Mulheisen M, Wu DK, Epstein DJ. Specification of the mammalian cochlea is dependent on Sonic hedgehog. Genes Dev. 2002;16:2365–2378. doi: 10.1101/gad.1013302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccomagno MM, Takada S, Epstein DJ. Wnt-dependent regulation of inner ear morphogenesis is balanced by the opposing and supporting roles of Shh. Genes Dev. 2005;19:1612–1623. doi: 10.1101/gad.1303905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha-Sanchez SM, Scheetz LR, Contreras M, Weston MD, Korte M, McGee J, Walsh EJ. Mature mice lacking Rbl2/p130 gene have supernumerary inner ear hair cells and supporting cells. J Neurosci. 2011;31:8883–8893. doi: 10.1523/JNEUROSCI.5821-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaghi M, Nasr M, Heller S. Concise review: inner ear stem cells–an oxymoron, but why? Stem Cells. 2012;30:69–74. doi: 10.1002/stem.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Brivanlou AH. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 2011;30:237–248. doi: 10.1038/emboj.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajan SA, Warchol ME, Lovett M. Toward a systems biology of mouse inner ear organogenesis: gene expression pathways, patterns and network analysis. Genetics. 2007;177:631–653. doi: 10.1534/genetics.107.078584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajan SA, Rubenstein JL, Warchol ME, Lovett M. Identification of direct downstream targets of Dlx5 during early inner ear development. Hum Mol Genet. 2011;20:1262–1273. doi: 10.1093/hmg/ddq567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, Smith J, van Heyningen V, Rubenstein JL, Livesey FJ. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5:e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner O, Eden A, Ben-Yosef T, Yanuka O, Simchen G, Benvenisty N. ECA39, a conserved gene regulated by c-Myc in mice, is involved in G1/S cell cycle regulation in yeast. Proc Natl Acad Sci USA. 1996;93:7143–7148. doi: 10.1073/pnas.93.14.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebe-Pedros A, de Mendoza A, Lang BF, Degnan BM, Ruiz-Trillo I. Unexpected repertoire of metazoan transcription factors in the unicellular holozoan Capsaspora owczarzaki. Mol Biol Evol. 2011;28:1241–1254. doi: 10.1093/molbev/msq309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipel K, Yanze N, Schmid V. Developmental and evolutionary aspects of the basic helix-loop-helix transcription factors Atonal-like 1 and Achaete-scute homolog 2 in the jellyfish. Dev Biol. 2004;269:331–345. doi: 10.1016/j.ydbio.2004.01.035. [DOI] [PubMed] [Google Scholar]
- Seo S, Lim JW, Yellajoshyula D, Chang LW, Kroll KL. Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. EMBO J. 2007;26:5093–5108. doi: 10.1038/sj.emboj.7601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata SB, Budenz CL, Bowling SA, Pfingst BE, Raphael Y. Nerve maintenance and regeneration in the damaged cochlea. Hear Res. 2010;281:56–64. doi: 10.1016/j.heares.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroyer NF, Helmrath MA, Wang VY, Antalffy B, Henning SJ, Zoghbi HY. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132:2478–2488. doi: 10.1053/j.gastro.2007.03.047. [DOI] [PubMed] [Google Scholar]
- Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, Degnan BM, Vervoort M. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol Biol. 2007;7:33. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simionato E, Kerner P, Dray N, Le Gouar M, Ledent V, Arendt D, Vervoort M. atonal- and achaete-scute-related genes in the annelid Platynereis dumerilii: insights into the evolution of neural basic-Helix-Loop-Helix genes. BMC Evol Biol. 2008;8:170. doi: 10.1186/1471-2148-8-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA, Fritzsch B, Pierce ML, Weston MD, Jahan I, McManus MT, Harfe BD. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev Biol. 2009;328:328–341. doi: 10.1016/j.ydbio.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staecker H, Praetorius M, Brough DE. Development of gene therapy for inner ear disease: using bilateral vestibular hypofunction as a vehicle for translational research. Hear Res. 2011;276:44–51. doi: 10.1016/j.heares.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulg M, Kirjavainen A, Pajusola K, Bueler H, Ylikoski J, Laiho M, Pirvola U. Differential sensitivity of the inner ear sensory cell populations to forced cell cycle re-entry and p53 induction. J Neurochem. 2010;112:1513–1526. doi: 10.1111/j.1471-4159.2009.06563.x. [DOI] [PubMed] [Google Scholar]
- Wagner A. The origins of evolutionary innovations. Oxford University Press; Oxford: 2011. [Google Scholar]
- Wang VY, Hassan BA, Bellen HJ, Zoghbi HY. Drosophila atonal fully rescues the phenotype of Math1 null mice: new functions evolve in new cellular contexts. Curr Biol. 2002;12:1611–1616. doi: 10.1016/s0960-9822(02)01144-2. [DOI] [PubMed] [Google Scholar]
- Weber T, Corbett MK, Chow LM, Valentine MB, Baker SJ, Zuo J. Rapid cell-cycle reentry and cell death after acute inactivation of the retinoblastoma gene product in postnatal cochlear hair cells. Proc Natl Acad Sci USA. 2008;105:781–785. doi: 10.1073/pnas.0708061105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Maklad A, Pirvola U, Fritzsch B. Brn3c null mutant mice show long-term, incomplete retention of some afferent inner ear innervation. BMC Neurosci. 2003;4:2. doi: 10.1186/1471-2202-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Yalamanchili HK, Li X, Yao KM, Sham PC, Zhang MQ, Wang J. Correlated evolution of transcription factors and their binding sites. Bioinformatics. 2011;27:2972–2978. doi: 10.1093/bioinformatics/btr503. [DOI] [PubMed] [Google Scholar]
- Yang J, Bouvron S, Lv P, Chi F, Yamoah EN. Functional features of trans-differentiated hair cells mediated by atoh1 reveals a primordial mechanism. J Neurosci. 2012;32:3712–3725. doi: 10.1523/JNEUROSCI.6093-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SL, Diolaiti D, Conacci-Sorrell M, Ruiz-Trillo I, Eisenman RN, King N. Premetazoan ancestry of the Myc-Max network. Mol Biol Evol. 2011;28:2961–2971. doi: 10.1093/molbev/msr132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Weber T, Yamashita T, Liu Z, Valentine MB, Cox BC, Zuo J. In vivo proliferation of postmitotic cochlear supporting cells by acute ablation of the retinoblastoma protein in neonatal mice. J Neurosci. 2010;30:5927–5936. doi: 10.1523/JNEUROSCI.5989-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JL, Gao WQ. Analysis of rat vestibular hair cell development and regeneration using calretinin as an early marker. J Neurosci. 1997;17:8270–8282. doi: 10.1523/JNEUROSCI.17-21-08270.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zine A, Aubert A, Qiu J, Therianos S, Guillemot F, Kageyama R, de Ribaupierre F. Hes1 and Hes5 activities are required for the normal development of the hair cells in the mammalian inner ear. J Neurosci. 2001;21:4712–4720. doi: 10.1523/JNEUROSCI.21-13-04712.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D, Erickson C, Kim EH, Jin D, Fritzsch B, Xu PX. Eya1 gene dosage critically affects the development of sensory epithelia in the mammalian inner ear. Hum Mol Genet. 2008;17:3340–3356. doi: 10.1093/hmg/ddn229. [DOI] [PMC free article] [PubMed] [Google Scholar]