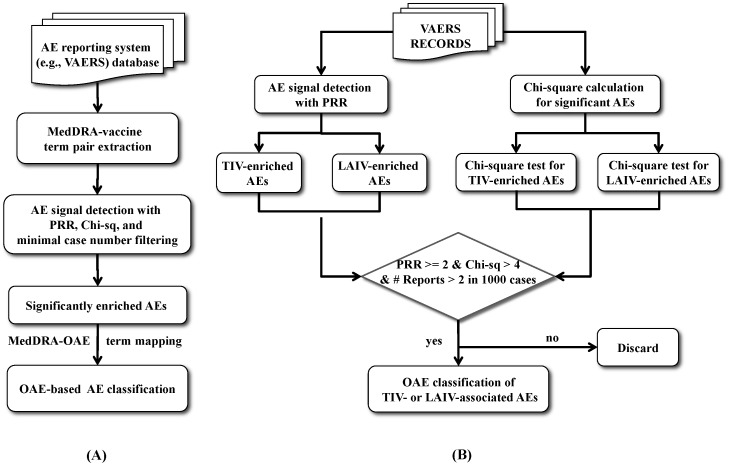
Figure 1. Workflow of the CODAE integrative AE bioinformatics analysis.
A generalized version of CODAE for detection of significant AE terms for one vaccine or one group of vaccines is outlined in (A). See details in the text. An expanded CODAE solution to analyze and compare AEs associated with the two vaccination groups is shown in (B). VAERS records were retrieved based on the query criteria of 4 TIVs (Afluria, Fluarix, Fluvirin, and Fluzone) year 1990–2011 and 1 LAIV (FluMist) year 2003–2011. Parallel analyses of the Proportional Reporting Ratios and Chi-square significant test were performed on individual AEs to identify enriched and significant AEs in each group. Base level filtration of 0.2% of total number of reports was also applied to each AEs. AEs that were identified to have PRR > = 2, Chi-square > = 4, and number of reports > = 0.2% of total reports were then classified based on OAE hierarchical structure. Classification of AEs filtered out AEs that overlapped between the 2 groups.