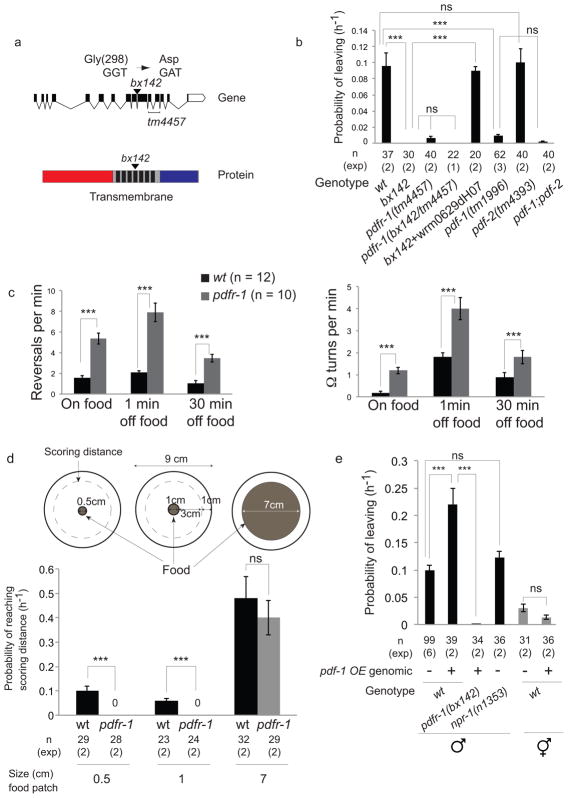
Figure 1. PDF-1 neuropeptide signaling stimulates mate searching.
(a) Genomic and protein domain structures of pdfr-1. (Top) Exons (black boxes) and introns (lines) of the pdfr-1 locus; (bottom) extracellular (red), 7 trans-membrane domains (black) and intracellular (blue) regions are shown. The changes in codon and amino acid produced by the bx142 mutation are indicated.
(b) Graph shows PL (probability of leaving food) values of wild type (wt), pdfr-1 (bx142) and (tm4457) mutants and mutants in the PDFR-1 ligands pdf-1(tm1996) and pdf-2(tm4393). The pdfr-1(tm4457) allele fails to complement bx142 indicating that the two mutations affect the same locus. The fosmid wrm0629dH07 contains the pdfr-1 locus. Error bars indicate SEM. n indicates total number of animals tested; (exp) indicates the number of independent population-based experiments. Maximum likelihood statistical analysis was used to compare PL values, ***p<0.001, ns: no statistically significant difference (p≥0.05).
(c) Graphs show the frequency of reversals and high angle turns (Ω turns) performed by wt and pdfr-1(bx142) mutant males during 5 minutes. Locomotion was observed on food and at two different time points off food: 1 minute after removal from food and 30 minutes after removal from food. n indicates total number of animals tested. Error bars indicate SEM. Statistical analysis was performed using Mann-Whitney test, ***p<0.001.
(d) Graphs show the rate at which wild-type (wt) and pdfr-1(bx142) males travel 3.5 cm, from the starting point at the center of the plate within a food patch to the scoring distance 1 cm away from the edge of the assay plate. This was calculated as in the leaving assay, as PL values (probability of reaching the scoring distance). Error bars indicate SEM. n indicates total number of animals tested; (exp) indicates the number of independent population-based experiments. Maximum likelihood statistical analysis was used to compare PL values (probability of reaching the scoring distance), ***p<0.001, ns: no statistically significant difference (p≥0.05).
(e) Graph shows PL values of wild-type and pdfr-1(bx142) mutant males and wild-type hermaphrodites with or without the pdf-1 genomic overexpression (pdf-1 OE) transgene. npr-1(n1353) mutant males display rates of mate-searching behavior similar to wild-type males. Error bars indicate SEM; n indicates total number of animals tested; (exp) indicates the number of independent population-based experiments. Maximum likelihood statistical analysis was used to compare PL values, ***p<0.001, ns: no statistically significant difference (p≥0.05).