ABSTRACT
Glucose-specific enzyme IIA (EIIAGlc) is a central regulator of bacterial metabolism and an intermediate in the phosphoenolpyruvate phosphotransferase system (PTS), a conserved phosphotransfer cascade that controls carbohydrate transport. We previously reported that EIIAGlc activates transcription of the genes required for Vibrio cholerae biofilm formation. While EIIAGlc modulates the function of many proteins through a direct interaction, none of the known regulatory binding partners of EIIAGlc activates biofilm formation. Therefore, we used tandem affinity purification (TAP) to compare binding partners of EIIAGlc in both planktonic and biofilm cells. A surprising number of novel EIIAGlc binding partners were identified predominantly under one condition or the other. Studies of planktonic cells revealed established partners of EIIAGlc, such as adenylate cyclase and glycerol kinase. In biofilms, MshH, a homolog of Escherichia coli CsrD, was found to be a dominant binding partner of EIIAGlc. Further studies revealed that MshH inhibits biofilm formation. This function was independent of the Carbon storage regulator (Csr) pathway and dependent on EIIAGlc. To explore the existence of multiprotein complexes centered on EIIAGlc, we also affinity purified the binding partners of adenylate cyclase from biofilm cells. In addition to EIIAGlc, this analysis yielded many of the same proteins that copurified with EIIAGlc. We hypothesize that EIIAGlc serves as a hub for multiprotein complexes and furthermore that these complexes may provide a mechanism for competitive and cooperative interactions between binding partners.
IMPORTANCE
EIIAGlc is a global regulator of microbial physiology that acts through direct interactions with other proteins. This work represents the first demonstration that the protein partners of EIIAGlc are distinct in the microbial biofilm. Furthermore, it provides the first evidence that EIIAGlc may exist in multiprotein complexes with its partners, setting the stage for an investigation of how the multiple partners of EIIAGlc influence one another. Last, it provides a connection between the phosphoenolpyruvate phosphotransferase (PTS) and Csr regulatory systems. This work increases our understanding of the complexity of regulation by EIIAGlc and provides a link between the PTS and Csr networks, two global regulatory cascades that influence microbial physiology.
Introduction
Vibrio cholerae is an intestinal pathogen and a natural inhabitant of aquatic environments (1). In these environments, V. cholerae is thought to exist in a surface-attached or biofilm state in or on zooplankton and insects, leading some to suggest that arthropods may serve as reservoirs or vectors of disease (2–8). Our laboratory has shown that V. cholerae forms a dense biofilm in the rectum of the model arthropod Drosophila melanogaster (9). Formation of this biofilm requires elaboration of a matrix comprised of the VPS exopolysaccharide as well as several structural proteins (10–16).
Transcription of the vps genes, which encode proteins required for biosynthesis of the V. cholerae biofilm matrix, is controlled by a complex regulatory network that integrates multiple environmental signals, including bacterial autoinducers, polyamines, nucleosides, indole, and carbohydrates transported by the phosphoenolpyruvate phosphotransferase system (PTS), a multicomponent phosphotransfer cascade (14, 17–22).
The PTS consists of four regulatory intermediates and a terminal apparatus that both transports and phosphorylates specific sugars (23). In order of phosphotransfer, the regulatory intermediates include enzyme I (EI), which accepts a phosphate from phosphoenolpyruvate (PEP), histidine protein (HPr), and enzymes IIA and IIB (EIIA and EIIB). Enzymes IIC, which are not part of the phosphotransfer cascade, form the transport apparatuses. While EI and HPr are considered to be general PTS components, each EII component responds to and transports a specific group of sugars. For this reason, the V. cholerae genome encodes 19 EIIA, -B, and -C homologs, each with distinct substrate specificities (24).
The phosphorylation state of PTS intermediates depends upon the intracellular pool of PEP and the abundance of PTS sugars in the environment. A high concentration of intracellular PEP increases the proportion of phosphorylated PTS intermediates, while transport of sugars through the PTS depletes the phosphate stored within the PTS. Bacteria are equipped with signal transduction pathways that monitor the phosphorylation state of PTS components and adjust their cellular physiology accordingly (25).
Our laboratory recently uncovered multiple independent pathways within the V. cholerae PTS that regulate synthesis of the biofilm matrix at the transcriptional level (26). One of these involves activation of biofilm formation by the glucose-specific EIIA component (EIIAGlc). The phosphorylated form of EIIAGlc modulates the action of the cAMP receptor protein (CRP), a global regulator of cellular physiology, by activating synthesis of cAMP through a direct interaction with adenylate cyclase (AC) (25). Because both CRP and cAMP have been reported to repress vps transcription and biofilm formation in V. cholerae (19, 27, 28), we hypothesized that EIIAGlc must interact with additional protein partners that activate biofilm formation. To test this, we isolated interaction partners of EIIAGlc in both planktonic and biofilm cells by tandem affinity purification (TAP) and then identified these proteins by mass spectrometry (29). Distinct EIIAGlc binding partners were identified in planktonic and biofilm cells. In particular, AC and glycerol kinase (GlpK), both well-established binding partners of EIIAGlc, were the most abundant interaction partners in planktonic cells, while MshH was the most abundant partner in the biofilm. We confirmed and further elucidated these interactions using a bacterial two-hybrid system. Both TAP and bacterial two-hybrid analyses suggested that factors in addition to the phosphorylation state of EIIAGlc contribute to the differential abundance of these protein-protein interactions in planktonic and biofilm cells.
MshH is a homolog of Escherichia coli CsrD, a component of an unusual and complex posttranscriptional regulatory cascade known as Carbon storage regulator (Csr) that has been uncovered by Romeo and colleagues in a series of elegant experiments (30–32). CsrD enhances the RNase E-dependent degradation of the small csr RNAs that inhibit binding of the regulatory protein CsrA to mRNA. Here we find that MshH represses V. cholerae biofilm formation only when EIIAGlc is present. While both MshH and EIIAGlc modulate the levels of the csr RNAs, this function does not play a role in regulation of biofilm formation in this V. cholerae strain. We conclude that MshH is a repressor of biofilm formation that may function upstream of EIIGlc. Furthermore, we hypothesize that EIIAGlc provides a platform for cooperative and competitive interactions between its many binding partners.
RESULTS
Tandem affinity purification reveals distinct EIIAGlc interaction partners in the planktonic and biofilm states.
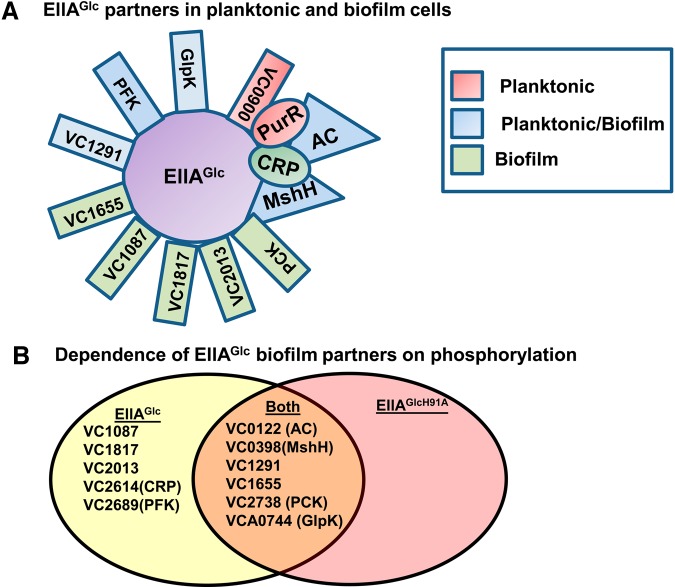
EIIAGlc regulates cellular physiology through interactions with other proteins. We used TAP followed by liquid chromatography-mass spectrometry (LC-MS) to identify interaction partners of EIIAGlc in planktonic and biofilm cells cultured in LB broth (Table 1 and Fig. 1A; see also Tables S1 and S2 in the supplemental material). Some proteins copurified with EIIAGlc in planktonic or biofilm cells specifically, while others copurified with EIIAGlc under both conditions (Fig. 1A; Table 1). Proving the validity of our analysis, glycerol kinase (GlpK) and AC copurified with EIIAGlc quite abundantly in planktonic cells. GlpK converts glycerol to glycerol-3-phosphate as it enters the cell. Unphosphorylated EIIAGlc is an established allosteric inhibitor of GlpK that blocks glycerol utilization when PTS substrates are present (33, 34). EIIAGlc-P has long been known to potentiate the activity of AC. However, direct contact between these two proteins has only recently been demonstrated (35). In this work, the investigators created an E. coli strain expressing native AC and another expressing AC tethered to an integral membrane protein. They found that EIIAGlc associated with the cell membrane only in cells expressing tethered AC. Our results build on these by showing that EIIAGlc interacts directly with the native form of AC.
TABLE 1 .
EIIAGlc and AC interaction partners
| Locus | Gene product | No. of specific peptidesb |
|||
|---|---|---|---|---|---|
| EIIAGlc mid-log |
EIIAGlc biofilm |
EIIAGlcH91A biofilm |
AC biofilm |
||
| VC0122a | Adenylate cyclase | 77 | 2 | 1 | (117) |
| VC0398 | MshH | 2 | 22 | 3 | 1 |
| VC0900 | GGDEF family protein | 6 | ND | ND | ND |
| VC0964 | EIIAGlc | (37) | (64) | (81) | 17 |
| VC1087 | Response regulator | ND | 1 | ND | ND |
| VC1291 | Hypothetical protein | 1 | 10 | 2 | 5 |
| VC1655 | Mg transporter | ND | 11 | 3 | ND |
| VC1721 | PurR | 7 | ND | ND | 2 |
| VC1817 | Fis family transcriptional regulator | ND | 1 | ND | ND |
| VC2013a | EIIBCGlc | ND | 1 | ND | ND |
| VC2614 | CRP | ND | 2 | ND | 6 |
| VC2689 | 6-Phosphofructokinase (PFK) | 4 | 2 | ND | 1 |
| VC2738 | Phosphoenolpyruvate carboxykinase (PCK) | ND | 8 | 10 | 1 |
| VCA0744a | Glycerol kinase (GlpK) | 100 | 2 | 6 | 4 |
Previously documented interaction with EIIAGlc.
Numbers indicate the total number of peptides identified that were specific to the indicated protein. “ND” indicates that peptides associated with this protein were not detected. Parentheses indicate identified peptides corresponding to the TAP-tagged protein.
FIG 1.
Analysis of the binding partners of EIIAGlc and adenylate cyclase in the biofilm state. (A) Diagram illustrating the binding partners of EIIAGlc found in planktonic cells, biofilm cells, or both. As proposed in the text, CRP and PurR are illustrated as binding to AC. (B) Venn diagram illustrating the overlap between the binding partners of EIIAGlcH91A and those of the wild-type EIIAGlc allele in biofilm cells.
Two proteins, PurR and the GGDEF domain-containing protein VC0900, copurified with EIIAGlc only in the planktonic state (Fig. 1A). Proteins copurifying with EIIAGlc in the biofilm alone included the putative response regulator VC1087, VC1655, a distant homolog of the cyclic nucleotide binding domain-containing magnesium transporter MgtE, VC1817, a GAF domain-containing σ54-dependent transcriptional regulator, EIIBCGlc, CRP, and PEP carboxykinase (PCK), which converts oxaloacetate to PEP. Proteins copurifying with EIIAGlc in both states included AC, GlpK, MshH, a predicted regulatory protein containing at least one cyclic nucleotide binding domain at locus VC1291, and 6-phosphofructokinase (PFK). PFK is an allosterically regulated enzyme in the glycolytic pathway that generates fructose 1,6-biphosphate. While AC and GlpK showed a strong preference for association with EIIAGlc in the planktonic state, MshH, VC1291, and PCK showed a strong preference for association in the biofilm state. Only PFK copurified equally well with EIIAGlc in both the planktonic and biofilm states.
Dependence of protein interactions in the biofilm on phosphorylation of EIIAGlc.
To investigate the dependence of biofilm-specific EIIAGlc binding partners on the phosphorylation state of EIIAGlc, we performed a biofilm TAP analysis using EIIAGlcH91A, a point mutant in which an alanine is substituted for the phosphorylated histidine. These results are listed in Table 1 (see also Table S3 in the supplemental material) and illustrated in Fig. 1B. Previous findings for E. coli suggest that GlpK interacts with the unphosphorylated form of EIIAGlc (36). However, in biofilm cells, it copurified only slightly better with EIIAGlcH91A than with native EIIAGlc. Of the proteins that interacted with EIIAGlc preferentially in the biofilm, some were able to interact with EIIAGlcH91A, while others were not. These data suggest that factors in addition to the phosphorylation state of EIIAGlc determine the specificity of these interactions for the biofilm state. However, it is also possible that the point mutation itself interferes with binding of some but not all of the interaction partners of EIIAGlc.
TAP analysis of AC binding partners suggests that EIIAGlc forms multiprotein complexes.
We were curious to know if EIIAGlc is able to interact concurrently with more than one binding partner in biofilm cells. To explore this further, we performed TAP analysis with AC and MshH, the principal binding partners of EIIAGlc in planktonic and biofilm cells, respectively. TAP analysis with MshH was not successful. The binding partners of AC revealed by TAP are shown in Table 1 and also in Table S4 in the supplemental material. EIIAGlc was the most abundant binding partner of AC in the biofilm. Furthermore, every other protein identified as a binding partner of AC had previously been identified as a binding partner of EIIAGlc. Some binding partners of EIIAGlc, including MshH, VC1655, and PCK, were poorly represented in the AC TAP analysis, while others, such as the transcription factors PurR and CRP, were better represented in the AC TAP analysis than in that with EIIAGlc. Our data suggest the hypotheses that (i) EIIAGlc and AC participate in multiprotein complexes with the other interaction partners identified here, (ii) PurR and CRP interact with AC, whereas the other partners interact with EIIAGlc, and (iii) the interactions of EIIAGlc with some protein partners are inhibited when AC is bound (Fig. 1A).
Bacterial two-hybrid assays further elucidate the interaction of EIIAGlc with AC and MshH.
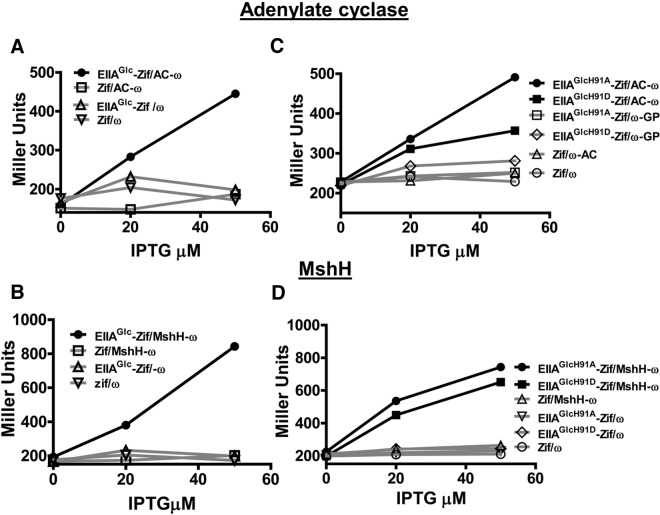
While our TAP analyses gave rise to many hypotheses, we chose to focus on AC and MshH, the dominant binding partners of EIIAGlc in planktonic and biofilm cells, respectively. To further examine the interaction of EIIAGlc with AC and MshH, bacterial two-hybrid assays were performed as follows. We constructed a plasmid encoding EIIAGlc fused to the N terminus of a zinc finger DNA-binding protein known as Zif and a plasmid encoding the putative EIIAGlc binding partner fused to the N terminus of the ω subunit of RNA polymerase. These plasmids were transformed into an E. coli strain carrying a Zif binding site upstream of the lacZ gene. In this strain, the level of β-galactosidase activity reflects the ability of EIIAGlc to recruit RNA polymerase to the lacZ promoter by interacting with its binding partner (37). Full-length adenylate cyclase was expressed for use in these assays. MshH has an N-terminal extracytoplasmic domain, a transmembrane domain, and C-terminal, cytoplasmic GGDEF and EAL domains. For these experiments, we expressed only the cytoplasmic portion of MshH, which consists of the terminal 578 amino acids of the protein. As shown in Fig. 2A and 2B, bacterial two-hybrid assays confirmed an interaction between EIIAGlc and both of these proteins. To examine the requirement for phosphorylation of EIIAGlc at position 91, we also performed two-hybrid assays with EIIAGlcH91A and EIIAGlcH91D, which mimic the unphosphorylated and phosphorylated forms of EIIAGlc, respectively. As shown in Fig. 2C and 2D, adenylate cyclase and MshH interacted well with both forms of EIIAGlc. Based on these findings and those of our TAP analyses, we hypothesize that factors in addition to the phosphorylation state of EIIAGlc determine the differential affinity of adenylate cyclase and MshH for EIIAGlc in planktonic and biofilm cells. For MshH, it is also possible that the extracellular or transmembrane portions of the protein, which were eliminated for these experiments, restrict its interaction with EIIAGlc to a particular phosphorylation state.
FIG 2.
Bacterial two-hybrid assays confirm the interaction of adenylate cyclase and MshH with native EIIAGlc and EIIAGlc point mutants mimicking the phosphorylated and unphosphorylated states. (A and B) Interaction of EIIAGlc fused to the N terminus of the DNA-binding protein Zif (EIIAGlc-Zif) with adenylate cyclase (AC-ω) (A) or MshH (MshH-ω) (B) fused to the ω subunit of RNA polymerase in an E. coli strain encoding the lacZ gene preceded by a Zif binding site. (C and D) A similar experiment was performed to assess the interactions of adenylate cyclase (AC-ω) (C) or MshH (MshH-ω) (D) with EIIAGlc point mutants in which an alanine (EIIAGlcH91A-Zif) or an aspartate (EIIAGlcH91D-Zif) was substituted for histidine 91 to mimic the unphosphorylated and phosphorylated states of EIIAGlc, respectively. Controls, which are traced in gray for each experiment, include (i) a vector encoding Zif alone (Zif) combined with a vector encoding the protein of interest fused to ω, (ii) a vector encoding native EIIAGlc or point mutants fused to Zif combined with a vector encoding ω alone (ω), and (iii) vectors encoding Zif and ω alone. Two experimental replicates were included in each trial, and several trials were performed. A representative trial is shown here.
MshH modulates biofilm formation in a Csr-independent manner.
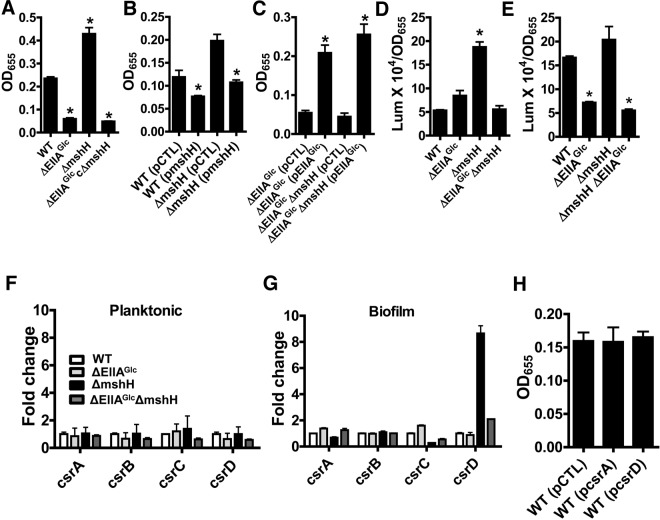
EIIAGlc is an activator of biofilm formation (26). Because MshH appeared to be the primary binding partner of EIIAGlc in the biofilm, we focused our subsequent studies on the role of MshH in regulation of biofilm formation. We first compared the biofilm-forming propensity of ΔEIIAGlc and ΔmshH mutants with that of wild-type V. cholerae. As shown in Fig. 3A, deletion of EIIAGlc decreased biofilm formation, while deletion of mshH increased biofilm formation. Because our findings proved that MshH was not the downstream factor responsible for activation of biofilm formation by EIIAGlc, we reasoned that MshH might interfere with potentiation of this downstream factor through its interaction with EIIAGlc. If this were the case, we predicted that deletion of mshH should have a biofilm phenotype only in the presence of EIIAGlc. To test this, we constructed a ΔmshH ΔEIIAGlc double mutant and measured its ability to form a biofilm. As predicted, MshH did not impact biofilm formation in the absence of EIIAGlc.
FIG 3.
Evidence that MshH inhibits activation of biofilm formation by EIIAGlc. (A) Biofilms formed by wild-type V. cholerae MO10 (WT) or ΔEIIAGlc, ΔmshH, and ΔEIIAGlc ΔmshH mutants. The biofilms formed by the ΔEIIAGlc (P < 0.0001), ΔmshH (P = 0.002), and ΔEIIAGlc ΔmshH (P < 0.0001) mutants are significantly different from that formed by wild-type V. cholerae. (B) Biofilm formation by a ΔmshH mutant carrying an empty vector (pCTL) or a plasmid encoding a wild-type allele of mshH (pmshH). The biofilms formed by the wild-type strain (P = 0.04) and the ΔmshH mutant (P = 0.003) carrying the plasmid expressing MshH are significantly different from those formed by the corresponding strains carrying a control plasmid. (C) Biofilm formed by ΔEIIAGlc or ΔEIIAGlc ΔmshH mutant carrying either an empty vector (pCTL) or a wild-type EIIAGlc allele (pEIIAGlc). The biofilm formed by the ΔEIIAGlc (pEIIAGlc) mutant is significantly different from that formed by the ΔEIIAGlc (pCTL) mutant (P = 0.0002). The biofilm formed by the ΔEIIA Glc ΔmshH (pEIIAGlc) mutant is significantly different from that formed by the ΔEIIA Glc ΔmshH (pCTL) mutant (P = 0.0002). (D and E) β-Galactosidase measurements reflecting planktonic (D) or biofilm (E) vpsL transcription in strains with chromosomal vpsL-lacZ reporter fusions. In planktonic cells, vpsL transcription in the ΔmshH mutant is significantly greater than that in wild-type cells (P = 0.0002). In the biofilm, vpsL transcription in the ΔEIIAGlc and ΔEIIAGlc ΔmshH strains is significantly different from that of wild-type V. cholerae (P < 0.0001). (F and G) qRT-PCR quantification of csrA, csrB, csrC, and csrD transcripts in planktonic and biofilm cultures. Transcript levels were measured in wild-type strains (WT) as well as ΔEIIAGlc, ΔmshH, and ΔEIIAGlc ΔmshH mutants. (H) Biofilm formation by wild-type V. cholerae with a control plasmid (pCTL), a plasmid expressing CsrA (pcsrA), or a plasmid carrying a csrD transcript (pcsrD).
To further confirm a role for MshH in regulation of biofilm formation, we cloned the wild-type mshH allele into an inducible vector and tested the effect of MshH overexpression on biofilm formation by both wild-type V. cholerae and a ΔmshH mutant. As shown in Fig. 3B, overexpression of MshH reduced biofilm formation in both wild-type and mutant cells. We further predicted that provision of EIIAGlc in trans would increase biofilm formation by the ΔmshH ΔEIIAGlc double mutant to levels equal to or greater than those observed for rescue of a ΔEIIAGlc mutant. As shown in Fig. 3C, this was in fact the case. Last, to examine the effect of these mutations on transcription of the vpsL genes, we performed β-galactosidase assays in planktonic (Fig. 3D) and biofilm (Fig. 3E) cells with our control V. cholerae strain as well as the ΔmshH, ΔEIIAGlc, and ΔmshH ΔEIIAGlc double mutants, which all have a vpsL-lacZ transcriptional fusion cloned into a neutral site on the chromosome. In planktonic cells, deletion of EIIAGlc had little effect on β-galactosidase activity, but deletion of mshH increased β-galactosidase activity 3-fold. When EIIAGlc was deleted in a ΔmshH mutant background, β-galactosidase activity decreased to that of the control strain. In biofilms, a different pattern was observed. Deletion of EIIAGlc decreased β-galactosidase activity, but deletion of MshH had only a small effect on β-galactosidase activity. Last, deletion of EIIAGlc in a ΔmshH mutant background reduced β-galactosidase activity to that of the ΔEIIAGlc mutant. These results suggest that both MshH and EIIAGlc regulate biofilm formation at the transcriptional level. MshH has its greatest effect on transcription of the vps genes in planktonic cells, while EIIAGlc has its greatest effect in biofilm cells. Because the effect of MshH on vps gene transcription in planktonic cells is dependent on EIIAGlc, we hypothesize that MshH interferes with activation of vps gene transcription by EIIAGlc in planktonic cells.
MshH is a homolog of E. coli CsrD, showing 33% identity and 53% similarity. CsrD accelerates degradation of the small RNAs csrB and csrC (32). These small RNAs, in turn, inhibit the action of CsrA, an mRNA-binding protein that regulates many aspects of cellular physiology at the posttranscriptional level (38). V. cholerae has three small RNAs that are similar to csrB and csrC. The third V. cholerae small RNA has also been called csrD. Therefore, to avoid confusion, we refer to the V. cholerae homolog of the CsrD protein as MshH.
We hypothesized that MshH and EIIAGlc might modulate the levels of the csr RNAs in V. cholerae. To measure the effect of MshH and EIIAGlc on levels of the csr RNAs, we quantified csrA, csrB, csrC, and csrD transcripts in both planktonic and biofilm cells by quantitative reverse transcriptase-PCR (qRT-PCR). As shown in Fig. 3F, deletion of either mshH or the EIIAGlc gene did not alter the levels of the csr RNAs in planktonic cells. In contrast, in biofilm cells, deletion of mshH increased levels of the csrD transcript approximately 8-fold (Fig. 3G). While deletion of EIIAGlc in a wild-type background had no effect on csr transcript levels, deletion of EIIAGlc in a ΔmshH mutant background decreased csrD levels in the biofilm. We hypothesize that while MshH modulates activation of the vps genes by EIIAGlc in planktonic cells, EIIAGlc modulates the role of MshH in degradation of csrD in the biofilm.
We predicted that if increased csrD transcript contributed to the biofilm phenotype of the ΔmshH mutant, overexpression of csrD should increase the biofilm formed by wild-type V. cholerae, and overexpression of csrA should decrease biofilm formation. As shown in Fig. 3H, neither of these interventions altered biofilm formation by wild-type V. cholerae. Therefore, we conclude that csrA and csrD do not modulate biofilm formation by this V. cholerae strain.
DISCUSSION
Previous experiments showed that EIIAGlc activates V. cholerae biofilm formation at the transcriptional level (26). We hypothesized that this occurred through an interaction with a novel signaling or regulatory protein. Therefore, we employed a proteomic approach to identify binding partners of EIIAGlc in planktonic and biofilm cells that might participate in regulation of biofilm formation. Our results suggested that the binding partners of EIIAGlc in planktonic and biofilm cells are distinct. In planktonic cells, we isolated known binding partners of EIIAGlc, such as adenylate cyclase and glycerol kinase. In addition, novel binding partners of EIIAGlc were identified in both planktonic and biofilm cells. These include a putative Mg2+ transporter, two additional metabolic enzymes, and several putative signaling proteins and transcription factors, including the V. cholerae CsrD homolog MshH. Through bacterial two-hybrid assays, we confirmed a direct interaction of MshH with EIIAGlc. Our data suggest that MshH inhibits V. cholerae biofilm formation by interfering with the ability of EIIAGlc to activate biofilm formation. Furthermore, although both MshH and EIIAGlc modulate cellular levels of the csr RNAs, these do not appear to be involved in regulation of biofilm formation. We propose a novel function for MshH as a regulatory link between the PTS and Csr global signaling pathways.
To our knowledge, this study represents the first comparison of the binding partners of EIIAGlc in planktonic and biofilm cells. In the past, the planktonic state has been used to study the binding partners of EIIAGlc. Therefore, it is not surprising that the known interaction partners of EIIAGlc, GlpK and AC, copurified most abundantly with EIIAGlc in the planktonic phase. In contrast, most of the biofilm-specific binding partners of EIIAGlc are novel. A common theme among these binding partners is the presence of conserved cyclic nucleotide binding domains, which are known to serve a regulatory function.
Several pieces of evidence suggest that the phosphorylation state of EIIAGlc alone does not account for the different interaction partners observed in planktonic and biofilm cells. First of all, both AC, which is thought to bind to phosphorylated EIIAGlc, and GlpK, which is thought to bind to unphosphorylated EIIAGlc, are major interaction partners in planktonic cells but not the biofilm. Second, only a subset of the proteins that interact with EIIAGlc in biofilm cells are able to interact with EIIAGlcH91A, which mimics the unphosphorylated form. Last, our bacterial two-hybrid studies conducted in E. coli suggest that MshH and AC interact well with point mutants that mimic both the unphosphorylated and phosphorylated forms of EIIAGlc. Therefore, we predict the existence of factors beyond phosphorylation state that determine the interactions of EIIAGlc with its protein partners in the biofilm and planktonic states.
We also studied the interaction partners of AC in biofilm cells. Based on these results, we hypothesize the existence of multiprotein complexes in biofilm cells that include EIIAGlc and AC. Because EIIAGlc is a small protein, binding partners that interact with it under similar conditions are likely to influence each other’s binding in either a cooperative or competitive manner.
mshH is the first gene in an operon encoding synthesis of the mannose-sensitive hemagglutinin pilus (MSHA), a structure that promotes attachment of single cells to surfaces but inhibits colonization of the mammalian intestine (39, 40). Previous research has shown that MshH is dispensable for pilus elaboration (41). The transcription factor ToxT coordinates repression of mshA and mshH with activation of the major virulence factors of V. cholerae, the toxin-coregulated pilus and cholera toxin (42). However, the significance of mshH regulation by ToxT requires further investigation.
MshH is comprised of an N-terminal periplasmic domain flanked by two predicted transmembrane domains. The cytoplasmic portion of MshH consists of a HAMP-like linker domain followed by GGDEF and EAL domains that are poorly conserved (43). In E. coli, these last two domains are not thought to play a direct role in modulating intracellular levels of c-di-GMP (32). Rather, they are thought to bind the csr small RNAs, which inhibit the Csr system, thus targeting them for degradation by RNase E. We found that mutation of MshH increases the csrD transcript in biofilm cells only. Interestingly, while mutation of EIIAGlc in a wild-type genetic background had no effect on csr transcript levels, deletion of EIIAGlc in a ΔmshH background returned the csrD transcript level almost to wild-type levels. One possible mechanism for this is that the EIIAGlc-MshH complex, which is found principally in the biofilm, is responsible for spatial colocalization of csrD and RNase E. In the absence of MshH, binding of csrD to EIIAGlc may protect it from degradation. However, it is also possible that MshH and EIIAGlc independently regulate csr RNAs at the transcriptional level. A direct measurement of RNA stability in the presence and absence of Msh and EIIAGlc would be required to distinguish between these two possibilities.
In V. cholerae, the CSR system inhibits the quorum-sensing master regulator HapR, resulting in activation of the vps genes and biofilm formation (44). In such strains, we predict that MshH would activate biofilm formation. However, disruption of the quorum-sensing cascade is frequently found in environmental and clinical strains of V. cholerae (45), and this is the case for the clinical strain used in the studies presented here. Therefore, our data are consistent with a model in which the csr RNAs do not affect biofilm formation when quorum sensing is disabled.
Many decades of investigation have established EIIAGlc as a regulator of nutrient uptake and utilization in planktonic bacteria (35, 36, 46, 47). Here we have shown that in V. cholerae, the biofilm-specific interactions of EIIAGlc are distinct from those in planktonic cells, suggesting that it participates in a different physiologic response. Furthermore, our findings suggest a novel paradigm for EIIAGlc as a hub for multiprotein complexes. Such complexes could provide a mechanism for interaction and competition among the binding partners of EIIAGlc.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used in this study are listed in Table S5 in the supplemental material. Bacteria were grown in LB. When required, the following antibiotics were added to the growth medium: streptomycin (100 µg/ml), ampicillin (50 or 100 µg/ml, as noted), carbenicillin (100 µg/ml), kanamycin (30 µg/ml), and tetracycline (10 µg/ml). For rescue experiments, protein expression was induced with 0, 0.01, 0.1, or 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). For bacterial two-hybrid assays, concentrations of 0, 20, and 50 µM IPTG were used for induction.
Tandem affinity purification and LC/MS analysis.
A bacterial strain encoding EIIAGlc with a C-terminal TAP tag at its native chromosomal location was generated as follows. A fragment encoding protein A linked by a tobacco etch virus (TEV) protease (TEVP) cleavage site to calmodulin binding protein was inserted into the suicide plasmid pGP704 between the SphI and SmaI restriction sites to create the plasmid pGP704-TAP (48). The terminal 382 bp of the EIIAGlc gene excluding the stop codon were amplified and ligated into pGP704-TAP in frame with the TAP sequence using SalI and SphI restriction sites. The plasmid was then inserted in the chromosome by homologous recombination. Correct insertion was verified by amplification of the inserted fragment from the chromosome and by Western analysis.
For purification of EIIAGlc interaction partners from mid-log, planktonic cells, V. cholerae strains were inoculated into 1-liter flasks containing LB supplemented with ampicillin and grown to an optical density at 655 nm (OD655) of 0.5. The planktonic cells were then pelleted, washed, resuspended in lysis buffer (20 mM K-HEPES [pH 7.9], 50 mM KCl, 0.5 mM dithiothreitol [DTT], and 10% glycerol) with protease inhibitors, and sonicated to disrupt cells. To isolate proteins interacting within the biofilm, static cultures of V. cholerae were grown for 60 h in 47-mm petri plates containing 40 ml of LB supplemented with ampicillin. The planktonic cells were aspirated, and the biofilm was washed three times with phosphate-buffered saline (PBS). The biofilm was then scraped off the plate into a 50-ml conical tube and washed in lysis buffer.
Particulates were removed from Trizol lysates by centrifugation, and clarified lysates were applied to a column containing either 400 µl (planktonic lysate) or 800 µl (biofilm lysate) of IgG Sepharose 6 Fast Flow resin (GE Healthcare) and incubated at 4°C for 2 h. The column was then washed three times with IPP150 buffer (10 mM Tris-HCl, 150 mM NaCl, and 0.1% NP-40) and equilibrated with TEVP cleavage buffer (IPP150 including 0.5 mM EDTA, 1 mM DTT, and 100 U TEV protease). This suspension was incubated overnight. After removal of the IgG resin, the supernatant was applied to a 400-µl volume of calmodulin affinity resin (Agilent Technologies). This was incubated for 1 h at 4°C, washed three times with IPP150 calmodulin binding buffer (10 mM β-mercaptoethanol, 10 mM Tris-HCl, pH 8, 150 mM NaCl, 1 mM magnesium acetate [MgOAc], 1 mM imidazole, 2 mM CaCl2, and 0.1% NP-40) and eluted in elution buffer (10 mM β-mercaptoethanol, 10 mM Tris-HCl, pH 8, 150 mM NaCl, 1 mM MgOAc, 1 mM imidazole, 2 mM EGTA, and 0.1% NP-40). Fractions containing the eluted proteins were concentrated using an Amicon Ultra-4 spin column and run into a 12% SDS-PAGE gel. Gel fragments above and below the tagged protein were removed and submitted for LC-MS analysis.
Generation of deletion mutants.
Gene deletions were engineered as previously described using the splice overlap extension (SOE) method and double homologous recombination (18, 49). The primers used for making deletions are listed in Table S6 in the supplemental material.
Construction of plasmids used for protein expression.
Rescue experiments were performed using pFLAG-CTC (Invitrogen). The pFLAG-MshH, pFLAG-csrD, and pFLAG-CsrA constructs were generated by PCR amplification of mshH, csrD, or csrA, respectively, using the primers listed in Table S6 in the supplemental material. These fragments were cloned into pFLAG–CTC.
Biofilm assays.
Vibrio cholerae strains to be tested were grown overnight and then diluted to an OD655 of 0.05 in fresh LB. Three hundred microliters of this cell suspension was aliquoted into three borosilicate tubes and incubated at 27°C for 21 to 24 h. Planktonic cells were separated from the biofilm, and planktonic growth was assessed by measuring the OD655. Three hundred microliters of PBS solution was added to the biofilm along with 1-mm-diameter glass beads (30% [vol/vol]) (BioSpec Products, Inc.). Tubes were vortexed to disperse biofilm-associated cells, and the OD655 of the resultant cell suspension was measured to quantify the number of cells associated with the biofilm. Error bars represent the standard deviation, and statistical significance was calculated using a Student t test.
Western blot analysis.
For detection of TAP-tagged proteins in elution fractions, 12 µl of each eluant fraction was added to 3 µl of 5× Laemmli buffer, boiled for 5 min, and centrifuged briefly. Protein components were separated on a 12% gel (Pierce) by SDS-PAGE. Proteins were transferred to a polyvinylidene difluoride (PVDF) membrane using a semidry transfer method. The membrane was then incubated for 1 h at room temperature in a blocking solution consisting of PBS with 0.1% Tween 20 (PBS-T) and 5% skim milk. Blocking solution was removed and replaced with fresh blocking solution supplemented with an horseradish peroxidase (HRP)-conjugated anti-calmodulin binding antibody (1:5,000) (Immunology Consultants Laboratory, Inc.). After incubation for 1 h, the membrane was washed three times with PBS-T and detected with ECL Plus Western blotting detection reagent (GE Healthcare).
Bacterial two-hybrid analysis.
For bacterial two-hybrid analysis, plasmids were constructed as follows. For construction of the pACTR-EIIAGlc-Zif plasmid, the gene encoding EIIAGlc was amplified and ligated into the plasmid pACTR-AP-Zif between NotI and BamHI restriction sites (37). Plasmids encoding the point mutants EIIAGlcH91A and EIIAGlcH91D were constructed similarly except that SOE was used to engineer point mutations prior to amplification. The plasmids pBRω-mshH and pBRω-cya were constructed by amplifying the genes encoding these proteins and cloning the resulting PCR products into the plasmid pBRGPω. Bacterial two-hybrid assays were performed as previously described (37). In brief, both vectors were electroporated into KDZif1∆Z cells. Transformants containing both vectors were selected on LB agar plates supplemented with kanamycin, carbenicillin, and tetracycline, inoculated into LB supplemented with the same antibiotics as well as either 0, 20, or 50 µM IPTG, and incubated overnight at 37°C with shaking. The following day, cultures were diluted in fresh LB supplemented with antibiotics and the indicated IPTG concentrations and grown to mid-log phase. Cells were harvested and used in β-galactosidase assays performed according to standard procedures. Two experimental replicates were performed in each trial, and several trials were performed.
β-Galactosidase assays.
Strains were grown overnight at 27°C on LB agar plates. Several colonies were resuspended in 1 ml of LB broth supplemented with streptomycin to yield the desired OD655. For mid-log assays, 5-ml cell suspensions of an OD655 of 0.004 to 0.005 were placed in 50-ml Falcon tubes and grown with agitation at 27°C for 3 h. Final OD655s, which were between 0.2 and 0.3, were recorded. These cells were pelleted, washed with 500 µl of Z buffer, and then resuspended in 250 µl of Z buffer. For biofilm assays, 300 µl of a 0.05-OD655 culture was grown statically for 18 to 20 h in 10- by 75-mm glass tubes (Fisher) at 27°C (50). The OD655 of the final culture was measured. A 150-µl volume of cells was pelleted and resuspended in 200 µl of Z buffer. For both planktonic and biofilm assays, cells were lysed by three freeze-thaw cycles, and 10 µl of the lysate was added to 70 µl of β-galactosidase Chemiluminescent Substrate Plus (Michigan Diagnostics, LLC) in a white 96-well plate and incubated at room temperature for 30 min. Triggering reagent (70 µl) was added to each well, and luminescence was read after 5 min. Three experimental replicates were included in each assay, and the assay was repeated multiple times. β-Galactosidase activity was reported as the luminescence measurement divided by the OD655 of the final culture.
Quantitative reverse transcription-PCR.
RNA was harvested either from mid-log cultures grown with aeration (planktonic cells) or from cultures grown statically at 27°C (biofilm cells). For planktonic cultures, cells were harvested at an OD655 of 0.5 and resuspended in 1 ml of Trizol (Invitrogen). For biofilm cultures, a mid-log culture of V. cholerae was inoculated into 90-mm petri dishes to yield a starting OD655 of 0.05, incubated for 19 h at 28°C, harvested into 50-ml conical tubes, resuspended in 2.5 ml of Trizol, and diluted 1:1 with fresh Trizol. RNA was isolated from 1 ml of the Trizol suspension as directed by the manufacturer. cDNA was synthesized using the SuperScript III kit (Invitrogen) as directed. Transcripts were quantified by qRT-PCR using the iTaq SYBR green kit (Bio-Rad) along with the Step One Plus real-time PCR system (Applied Biosystems). Transcript levels of csrA, csrB, csrC, and csrD were normalized to that of clpX. Fold expression was determined using the ΔΔCT method. Three experimental replicates were performed in each trial, and multiple trials were performed.
SUPPLEMENTAL MATERIAL
Mid-log tandem affinity purification results with EIIAGlc.
Biofilm tandem affinity purification results with EIIAGlc.
Biofilm tandem affinity purification results with EIIAGlcH91A.
Biofilm tandem affinity purification results with AC.
Strains and plasmids.
Primers.
ACKNOWLEDGMENTS
We thank Zhipeng Wang, Simon Dove, Kirsty McFarland, and Josh Sharp for helpful discussions and guidance with tandem affinity purifications and bacterial two-hybrid assays. We also thank Tony Romeo and Laetitia Houot for insightful suggestions. Finally, we thank Krista Pickering for her support and understanding during the course of this work. Proteomic analysis was conducted at the Taplin proteomics core at Harvard Medical School with expert assistance from Ross Tomaino.
This work was supported by AHA 09GRNT2280096 and NIH AI050032 to P.I.W.
Footnotes
Citation Pickering BS, Smith DR, Watnick PI. 2012. Glucose-specific enzyme IIA has unique binding partners in the Vibrio cholerae biofilm. mBio 3(6):e00228-12. doi:10.1128/mBio.00228-12.
REFERENCES
- 1. Colwell RR, Spira WM. 1992. The ecology of Vibrio cholerae, p 107–127 In Barua D, Greenough WB, III, Cholera. Plenum, New York, NY. [Google Scholar]
- 2. Abdel-Monem MH. 1988. House flies (Musca domestica) as a vector of some enteric pathogens in an Egyptian village. J. Egypt. Public Health Assoc. 63:199–208 [PubMed] [Google Scholar]
- 3. Broza M, Gancz H, Halpern M, Kashi Y. 2005. Adult non-biting midges: possible windborne carriers of Vibrio cholerae non-O1 non-O139. Environ. Microbiol. 7:576–585 [DOI] [PubMed] [Google Scholar]
- 4. Echeverria P, Harrison BA, Tirapat C, McFarland A. 1983. Flies as a source of enteric pathogens in a rural village in Thailand. Appl. Environ. Microbiol. 46:32–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huq A, et al. 1996. A simple filtration method to remove plankton-associated Vibrio cholerae in raw water supplies in developing countries. Appl. Environ. Microbiol. 62:2508–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Khin Nwe O, Sebastian AA, Aye T. 1989. Carriage of enteric bacterial pathogens by house flies in Yangon, Myanmar. J. Diarrhoeal Dis. Res. 7:81–84 [PubMed] [Google Scholar]
- 7. Sack RB, et al. 2003. A 4-year study of the epidemiology of Vibrio cholerae in four rural areas of Bangladesh. J. Infect. Dis. 187:96–101 [DOI] [PubMed] [Google Scholar]
- 8. Sukontason K, et al. 2000. Mechanical carrier of bacterial enteric pathogens by chrysomya megacephala (Diptera: Calliphoridae) in Chiang Mai, Thailand. Southeast Asian J. Trop. Med. Public Health 31(Suppl 1):157–161 [PubMed] [Google Scholar]
- 9. Purdy AE, Watnick PI. 2011. Spatially selective colonization of the arthropod intestine through activation of Vibrio cholerae biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 108:19737–19742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Absalon C, Van Dellen K, Watnick PI. 2011. A communal bacterial adhesin anchors biofilm and bystander cells to surfaces. PLoS Pathog. 7:e1002210 http://dx.doi.org/10.1371/journal.ppat.1002210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Berk V, et al. 2012. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science 337:236–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fong JC, Karplus K, Schoolnik GK, Yildiz FH. 2006. Identification and characterization of RbmA, a novel protein required for the development of rugose colony morphology and biofilm structure in Vibrio cholerae. J. Bacteriol. 188:1049–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fong JC, Yildiz FH. 2007. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J. Bacteriol. 189:2319–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moorthy S, Watnick PI. 2005. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol. Microbiol. 57:1623–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Watnick PI, Kolter R. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yildiz FH, Schoolnik GK. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 96:4028–4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hammer BK, Bassler BL. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101–104 [DOI] [PubMed] [Google Scholar]
- 18. Haugo AJ, Watnick PI. 2002. Vibrio cholerae CytR is a repressor of biofilm development. Mol. Microbiol. 45:471–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Houot L, Watnick PI. 2008. A novel role for enzyme I of the Vibrio cholerae phosphoenolpyruvate phosphotransferase system in regulation of growth in a biofilm. J. Bacteriol. 190:311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Karatan E, Duncan TR, Watnick PI. 2005. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J. Bacteriol. 187:7434–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McGinnis MW, et al. 2009. Spermidine regulates Vibrio cholerae biofilm formation via transport and signaling pathways. FEMS Microbiol. Lett. 299:166–174 [DOI] [PubMed] [Google Scholar]
- 22. Mueller RS, Beyhan S, Saini SG, Yildiz FH, Bartlett DH. 2009. Indole acts as an extracellular cue regulating gene expression in Vibrio cholerae. J. Bacteriol. 191:3504–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Postma PW, Lengeler JW, Jacobson GR. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Houot L, Chang S, Absalon C, Watnick PI. 2010. Vibrio cholerae phosphoenolpyruvate phosphotransferase system control of carbohydrate transport, biofilm formation, and colonization of the germfree mouse intestine. Infect. Immun. 78:1482–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lengeler JW, Jahreis K. 2009. Bacterial PEP-dependent carbohydrate: phosphotransferase systems couple sensing and global control mechanisms. Contrib. Microbiol. 16:65–87 [DOI] [PubMed] [Google Scholar]
- 26. Houot L, Chang S, Pickering BS, Absalon C, Watnick PI. 2010. The phosphoenolpyruvate phosphotransferase system regulates Vibrio cholerae biofilm formation through multiple independent pathways. J. Bacteriol. 192:3055–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fong JC, Yildiz FH. 2008. Interplay between cyclic AMP-cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholerae biofilm formation. J. Bacteriol. 190:6646–6659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liang W, Silva AJ, Benitez JA. 2007. The cyclic AMP receptor protein modulates colonial morphology in Vibrio cholerae. Appl. Environ. Microbiol. 73:7482–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Puig O, et al. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218–229 [DOI] [PubMed] [Google Scholar]
- 30. Liu MY, Romeo T. 1997. The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. J. Bacteriol. 179:4639–4642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Romeo T. 1998. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 29:1321–1330 [DOI] [PubMed] [Google Scholar]
- 32. Suzuki K, Babitzke P, Kushner SR, Romeo T. 2006. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 20:2605–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Feese M, Pettigrew DW, Meadow ND, Roseman S, Remington SJ. 1994. Cation-promoted association of a regulatory and target protein is controlled by protein phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 91:3544–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van der Vlag J, van Dam K, Postma PW. 1994. Quantification of the regulation of glycerol and maltose metabolism by IIAGlc of the phosphoenolpyruvate-dependent glucose phosphotransferase system in Salmonella typhimurium. J. Bacteriol. 176:3518–3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Park YH, Lee BR, Seok YJ, Peterkofsky A. 2006. In vitro reconstitution of catabolite repression in Escherichia coli. J. Biol. Chem. 281:6448–6454 [DOI] [PubMed] [Google Scholar]
- 36. Hurley JH, et al. 1993. Structure of the regulatory complex of Escherichia coli IIIGlc with glycerol kinase. Science 259:673–677 [PubMed] [Google Scholar]
- 37. Charity JC, et al. 2007. Twin RNA polymerase-associated proteins control virulence gene expression in Francisella tularensis. PLoS Pathog. 3:e84 http://dx.doi.org/10.1371/journal.ppat.0030084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Timmermans J, Van Melderen L. 2010. Post-transcriptional global regulation by CsrA in bacteria. Cell. Mol. Life Sci. 67:2897–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hsiao A, Liu Z, Joelsson A, Zhu J. 2006. Vibrio cholerae virulence regulator-coordinated evasion of host immunity. Proc. Natl. Acad. Sci. U. S. A. 103:14542–14547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moorthy S, Watnick PI. 2004. Genetic evidence that the Vibrio cholerae monolayer is a distinct stage in biofilm development. Mol. Microbiol. 52:573–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marsh JW, Taylor RK. 1999. Genetic and transcriptional analyses of the Vibrio cholerae mannose-sensitive hemagglutinin type 4 pilus gene locus. J. Bacteriol. 181:1110–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hsiao A, Xu X, Kan B, Kulkarni RV, Zhu J. 2009. Direct regulation by the Vibrio cholerae regulator ToxT to modulate colonization and anticolonization pilus expression. Infect. Immun. 77:1383–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schirmer T, Jenal U. 2009. Structural and mechanistic determinants of c-di-GMP signalling. Nat. Rev. Microbiol. 7:724–735 [DOI] [PubMed] [Google Scholar]
- 44. Lenz DH, Miller MB, Zhu J, Kulkarni RV, Bassler BL. 2005. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol. Microbiol. 58:1186–1202 [DOI] [PubMed] [Google Scholar]
- 45. Joelsson A, Liu Z, Zhu J. 2006. Genetic and phenotypic diversity of quorum-sensing systems in clinical and environmental isolates of Vibrio cholerae. Infect. Immun. 74:1141–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saier MH, Jr, Roseman S. 1976. Sugar transport. 2nducer exclusion and regulation of the melibiose, maltose, glycerol, and lactose transport systems by the phosphoenolpyruvate:sugar phosphotransferase system. J. Biol. Chem. 251:6606–6615 [PubMed] [Google Scholar]
- 47. Saier MH, Jr, Roseman S. 1976. Sugar transport. The crr mutation: its effect on repression of enzyme synthesis. J. Biol. Chem. 251:6598–6605 [PubMed] [Google Scholar]
- 48. Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Horton RM, Cai ZL, Ho SN, Pease LR. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8:528–535 [PubMed] [Google Scholar]
- 50. Miller JH. 1992. A Short Course in Bacterial Genetics. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mid-log tandem affinity purification results with EIIAGlc.
Biofilm tandem affinity purification results with EIIAGlc.
Biofilm tandem affinity purification results with EIIAGlcH91A.
Biofilm tandem affinity purification results with AC.
Strains and plasmids.
Primers.