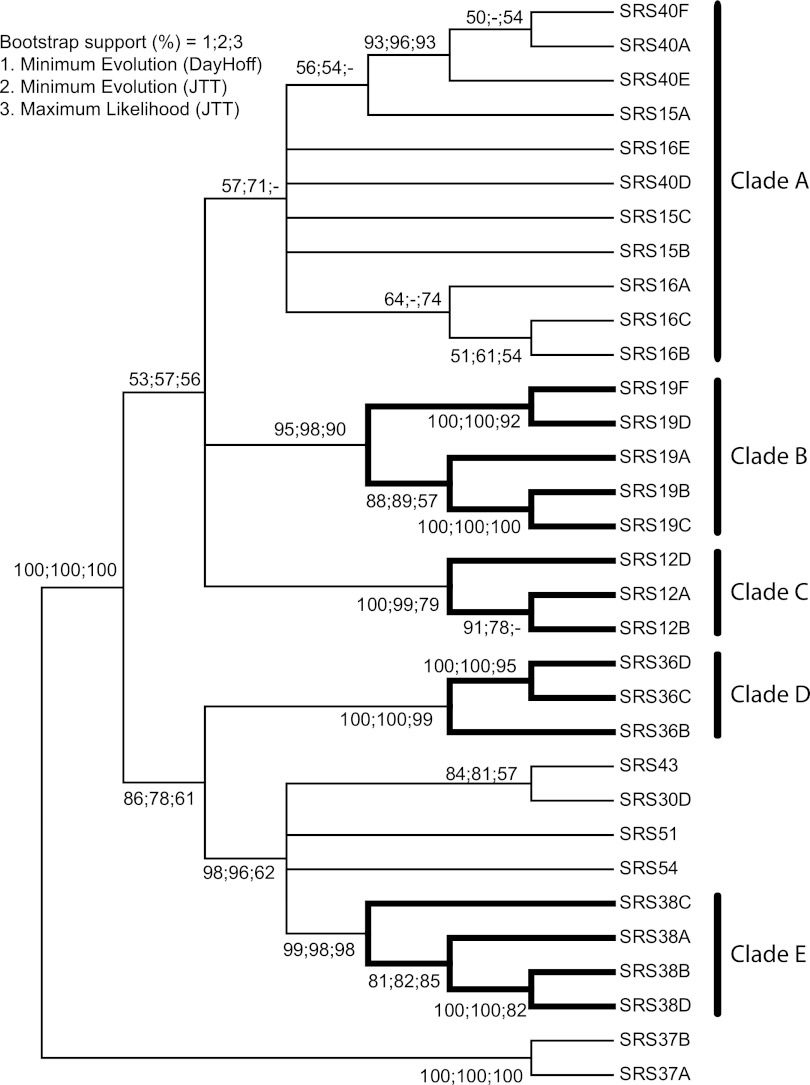
FIG 4.
Phylogenetic relationship of the most common SRS domain architectures. The alignment and tree reconstructions were performed as described in Materials and Methods. To achieve a robust phylogeny, three tree-building methods were used, i.e., ME with DayHoff and JTT probability models and ML with the JTT model. The three methods returned highly similar topologies. The node supports shown are from 1,000 bootstrap replicates. Nodes are considered resolved if they gained >50% support in two of the three tree-building methods used. Only proteins containing nondegraded domains were included in the analysis