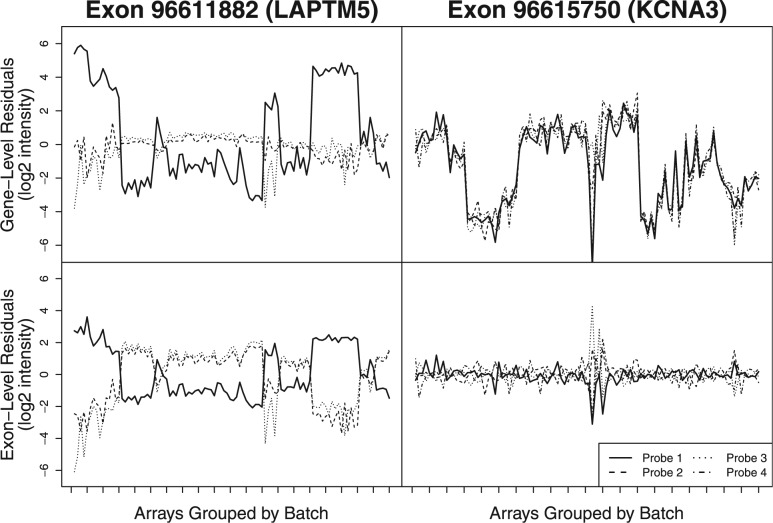
Fig. 1.
Residuals for probes targeting one of two exons are shown after fitting a standard RMA model to 100 arrays from 20 different batches (unique experiment/tissue combinations) at both gene (upper panels) and exon levels (lower panels). For both exons, Probe 1 (solid black line) appears to have a strong batch effect (high between-batch residual variance) when assessing probes at the gene level. However, in the case of Exon 96 615 750, the other three probes targeting this exon have nearly the same pattern of residuals across batches. This suggests that the high residual variance may be due to alternative splicing rather than a batch effect. By assessing probes at the exon level (lower panels), one still observes the high between-batch residual variance seen for Probe 1 targeting Exon 96 611 882 (left), but not for the probes targeting Exon 96 615 750 (right). By evaluating probe behavior at the exon level, we are able to distinguish between batch effects and splice variants