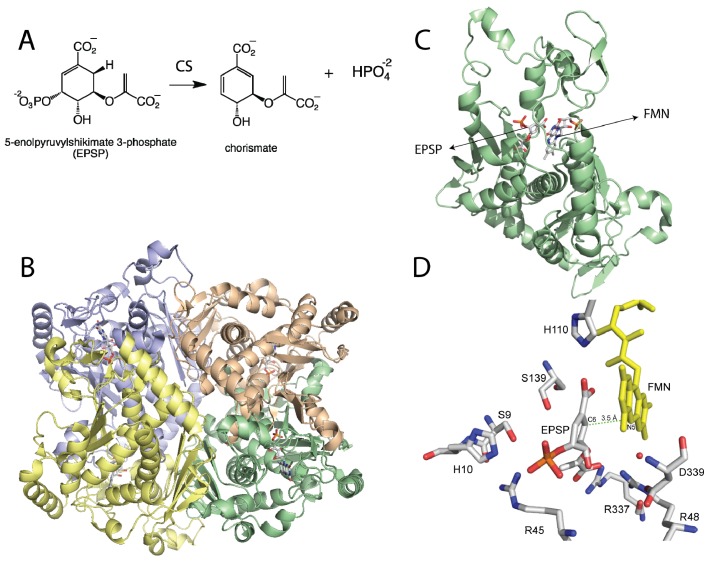
Figure 1.
(A) Reaction catalyzed by chorismate synthase; (B) Homotetrameric structure of Chorismate synthase (CS) from Streptococcus pneumoniae (PDB code 1QXO), where each monomer is shown in a different color; (C) Monomer of CS showing the flavin mononucleotide (FMN) and with 5-enolpyruvylshikimate 3-phosphate (EPSP); (D) Active site of CS with EPSP bound. The N5-position of the flavin is at the proper distance and orientation to act as the active site base. Residues that have been shown to be important in catalysis are shown. D339 is predicted to activate a water molecule (red sphere) for deprotonation of the N5-position of the FMN. H110 is close to the N1-O1 locus, where it can donate a proton to form the neutral reduced flavin as proposed. The other residues are involved in substrate binding or proton donation during phosphate release [26–28].