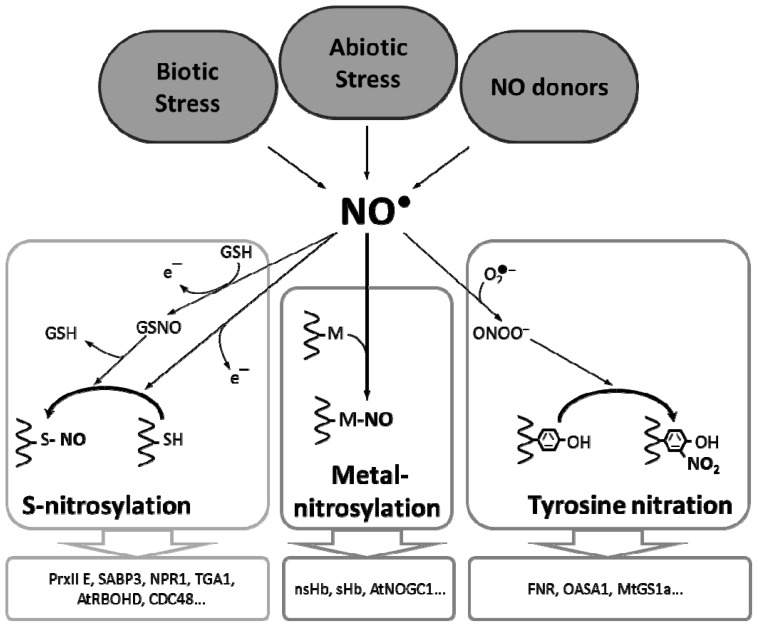
Figure 1.
Schematic illustration of NO dependent PTM in plants. To date, all the analyses of NO-modified proteins in plants followed an NO production induced by (a)biotic stress or NO donors treatment. The NO radical can react with transition metals (M) of metalloproteins. This process is called metal nitrosylation and can affect notably (non)-symbiotic hemoglobins (nsHb and sHb) and an Arabidopsis thaliana NO-dependent guanylate cyclase (AtNOGC1). The Tyr nitration depends on the formation of NO derivatives, particularly peroxynitrite formed in the presence of the superoxide anion (O2•−). Nitration occurs on one of the two carbon equivalent (C3) of the aromatic ring of tyrosine residues to form a 3-nitrotyrosine. This reaction has been demonstrated in plants for the ferredoxin-NADP oxidoreductase (FNR), the guanylate cyclase of Medicago truncatula (MtGS1a) or the of O-acetylserine(thiol)lyase A1 (OASA1). Protein S-nitrosylation is the electrophilic attack of nitrosonium cation (NO+, resulting from the oxidation of NO) on a thiolate group of a cysteine residue of a target protein. Among numerous proteins, this posttranslational modification affect for example peroxyredoxin II E (PrxII E), salicylic acid binding protein 3 (SABP3), nonexpressor of pathogenesis-related gene 1 (NPR1), transcription factor TGA1, respiratory burst oxidase homologue D (RBOHD) or cell division cycle 48 (CDC48). All these modifications will participate to the change of the plant cell physiology depending on the stimulus applied.