Figure 1.
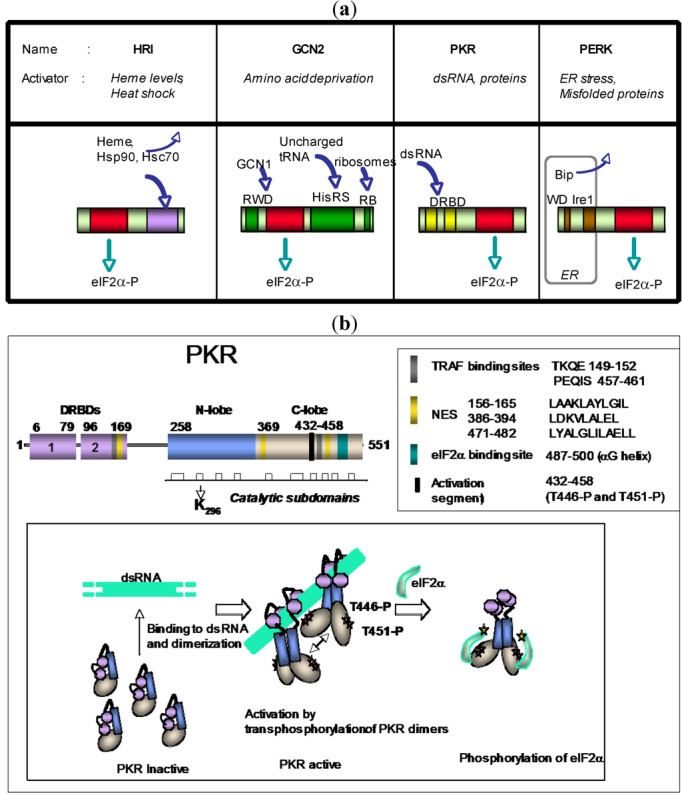
(a) The eIF2α-kinase family. Schematic representation of the four eIF2α-kinases: HRI, GCN2, PKR and PERK, showing their catalytic domains (in red) located at the N terminus (HRI, GCN2) or C terminus (PKR and PERK). HRI activates in response to diminution of heme levels or binding to some heat shock proteins such as Hsp90 or Hsc70. GCN2 form an inactive dimer through its region homologous to histidyl-tRNA synthetase (HisRS) and becomes activated, by change of conformation, when this domain binds uncharged tRNA, allowing its N-terminal ring finger and WD repeat domain (RWD) to bind GCN1 required for its function. It contains also a Ribosomal Binding site (RB) at its C terminus. PKR activates through two basic domains or dsRNA binding domains (DRBD) located at its N terminus. PERK is maintained in an inactive state through binding to the chaperone Bip protein (also known as Grp78 or Hsp 70-5) at its luminal N terminus (on WD repeats and Ire1-like domain). Unfolded proteins appearing in the lumen during ER stress attract Bip, which allows PERK homodimerization and activation of its cytosolic kinase domain. (b) PKR structure and mode of activation. The two 73 aa dsRNA Binding Domains (DRBD1 and DRBD2), located at the N terminus of PKR (purple) are responsible for the binding of PKR to its regulators. Mutation of the K296 residue (in subdomain II of the 253–525 catalytic domain; white bars) is sufficient to abrogate the catalytic activity of PKR. The sequences of the TRAF binding sites (grey bars) and nuclear export sequences (NES; yellow bars) is as indicated in the upper box. The eIF2α substrate binds in the helical C-lobe of PKR (blue bar) and the activation segment (black) contains the threonines 446 and 451. Lower box: Representation of the activation process of PKR upon binding to dsRNA. After binding of the first DRBD to dsRNA, binding of the second DRBD stabilizes the dsRNA/PKR complex and opens the conformation of PKR. This allows PKR dimerization through the N-lobe of its kinase domain (blue). PKR dimers activate each other by transphosphorylation of their T446 and T551 residues (red stars). The substrate (here eIF2α; green) can then access to its docking site and is positioned correctly within the catalytic domain of PKR where its acceptor site (yellow star) can receive the phosphate from ATP.
