Abstract
Heteromorphic sex chromosomes, where one sex has two different types of sex chromosomes, face very different evolutionary consequences than do the autosomes. Two important features of sex chromosomes arise from being present in only copy in one of the sexes: dosage compensation and the meiotic silencing of sex chromosomes. Other differences arise because sex chromosomes spend unequal amounts of time in each sex. Thus, the impact of evolutionary processes (mutation, selection, genetic drift, and meiotic drive) differs substantially between each sex chromosome, and between the sex chromosomes and the autosomes. Sex chromosomes also play a disproportionate role in Haldane’s rule and other important patterns related to hybrid incompatibility, and thus speciation. We review the consequences of sex chromosomes on hybrid incompatibility. A theme running through this review is that epigenetic processes, notably those related to chromatin, may be more important to the evolution of sex chromosomes and the evolution of hybrid incompatibility than previously recognized.
Keywords: sex chromosomes, hybrid incompatibility, speciation, evolution
Why study the evolutionary genetics of sex chromosomes?
Diverse taxa of multicultural eukaryotes (e.g., plants, insects, fish, mammals, birds) have dimorphic sex chromosomes. In such cases, chromosomes that look different from one another and from the autosomes (nonsex chromosomes) determine sex.1–3 Sex chromosome systems vary considerably. In some organisms, as in mammals and many insects, the males are heterogametic (have two different sex chromosomes) and females are homogametic (have two of the same sex chromosome). In others, such as birds and butterflies, females are heterogametic and males are homogametic. Following standard notation, females and males are XX and XY, respectively, in male heterogametic systems, and males and females are ZZ and ZW, respectively, in female heterogametic systems. Both male heterogamety and female heterogamety have independently evolved numerous times.1
This review focuses on the consequences of having heteromorphic sex chromosomes. Two major consequences arise because the heterogametic sex has only a single copy of each sex chromosome: the need to equalize the dosage of the expression of genes between the sexes and between the sex chromosomes and the autosomes, and the failure of sex chromosomes to pair in meiosis in the heterogametic sex. Other consequences arise because sex chromosomes experience environments differently from each other and from the autosomes because they are unequally represented in the sexes. These differences have implications for several major evolutionary genetic processes (mutation, selection, random genetic drift, recombination, and genomic conflict). In the following sections of this review, we provide a detailed exploration of the multifarious consequences, focusing on recent studies.
We then turn to the evolutionary origins and dynamics and of sex chromosome evolution. Because this subject has been the topic of several excellent recent reviews (e.g., 1–3), we will not provide comprehensive coverage of that material, but instead will bring up salient points and recent studies, considering both male and female heterogametic systems. We then review recent studies that find genes that are more strongly expressed in one sex than the other tend to be found on sex chromosomes much more or much less often than expected.
The last, and largest, part of this review concerns the consequences of sex chromosome evolution to hybrid incompatibility, the severe reduction in fitness of hybrids between species. Sex chromosomes are central to hybrid incompatibility4,5 as illustrated by the strong links between sex chromosomes and three major patterns of hybrid incompatibility: Haldane’s rule, the large X effect, and the asymmetry of hybrid viability and fertility in reciprocal crosses. Haldane’s rule comes from an observation Haldane6 made: in crosses between two species when one sex of the F1 hybrids had a lower viability or fertility than the other, the heterogametic sex was more affected than the homogametic sex. This empirical generalization remains the focus of much work in speciation genetics.7–13 The large X effect is the finding that the X chromosome appears to have a disproportionately larger effect on hybrid incompatibility than expected based on its size.4,7 Recent genetic studies in the genus Drosophila demonstrate that the large X effect is due to a greater density of hybrid incompatibility genetic factors.14–16 An analogous large Z effect has been found in butterflies.17,18 Finally, the viabilities and fertilities of hybrids from reciprocal crosses often differ. This asymmetry, dubbed Darwin’s corollary to Haldane’s rule,19 frequently arises due to the large X effect.
Recent reviews of hybrid incompatibility have suggested that genomic conflict is a major driver of the evolution of hybrid incompatibility.20–23 Sex chromosomes, due to their asymmetric mode of inheritance, are disproportionately likely to be involved in drive (e.g., Refs. 20, 21, 24–26). Thus, conflict may explain, at least in part, why the sex chromosomes play a disproportionate role in the evolution of hybrid incompatibility. The linkage of these three important speciation patterns (Haldane’s rule, the large X effect, and asymmetric effects in reciprocal crosses) to sex chromosomes and the connections between sex chromosomes, genomic conflict, and hybrid incompatibility should prompt those interested in the evolutionary genetics of hybrid incompatibility to consider the effects of sex chromosomes.
What are the mechanistic consequences of hemizygous sex chromosomes?
A salient feature of sex chromosomes is that they are present in only a single copy in the heterogametic sex. In many organisms, one sex chromosome (usually the Y or the W) is missing a large proportion of the genes of the other. Thus, the heterogametic sex can be hemizygous for a substantial proportion of the genome. Several consequences stem from this hemizygosity. One is dosage compensation, which has two parts: equalizing the gene dosage of the X (Z) with those of autosomal genes and equalizing the gene dosage of X (Z) linked genes between males and females. Another is the silencing of the sex chromosomes during meiosis (meiotic sex chromosome inactivation, MSCI).
Dosage compensation is achieved in a myriad of ways in different taxa.27 In eutherian and marsupial mammals, a large part of one of the X chromosomes in females is silenced, and thus, both males and females have but a single active X chromosome.28 Eutherian mammals and marsupials differ with respect to which X chromosome is silenced, with the paternal X being silenced in marsupials and a random X being silenced in eutherian mammals.29,30 In contrast, dosage compensation in Drosophila involves the hyperactivation of the X chromosome in males.31
The biochemical mechanisms of Drosophila dosage compensation have been worked out.27,32 A key feature is the recruitment of a protein/RNA complex known as MSL (male-specific lethal) to the X. The presence of this complex causes the aceylation of histone H4, which changes the chromatin structure, and results in an elevated transcription rate of X-linked genes.33 MSL first binds to high-affinity sites along the X, with the result that genes nearer the high-affinity sites are more likely to be affected by this process than genes further away.34
New research and novel datasets have spurred debate as to the scope of dosage compensation between the X and the autosomes.35–37 A recent studyin mammals using RNAseq suggested that the ratio of expression of X-linked genes to autosomes is around 0.5, and not 1.0 as would be expected under dosage compensation between autosomal and X-linked genes.36 In nematodes, RNAseq data show that X-linked and autosomal genes have comparable expression levels in larvae, but that the X-linked genes have roughly half the expression of autosomal genes in adults.36 Thus, nematode dosage compensation appears to be transient.35,36 However, experiments that take into account the skewed gene content of X-linked genes (biased toward reproductive function and germline expression) indicate that that X-linked expression is compensated in mammals, C. elegans, and D. melanogaster.37
Another consequence of hemizygosity of the sex chromosomes is that large parts of heteromorphic chromosomes cannot pair normally in meiosis. This lack of pairing is believed to promote MSCI, the silencing of sex chromosomes that begins in the pachytene stage of meiosis. MSCI, which has been best characterized in mammals,38 is an epigenetic phenomenon that involves chromatin remodeling (as well as DNA repair, specifically the repair after double-stranded breaks that occur in meiotic recombination).38 MSCI is actually “a special example of a more general mechanism called meiotic silencing of unsynapsed chromatin (MSUC), which silences chromosomes that fail to pair with their homologous partners and, in doing so, may protect against aneuploidy in subsequent generations” (see Ref. 38, p. 1823). Failure of proper MSCI has been linked to infertility in both mice and humans.38
Recent studies have characterized MSCI in opossums39 and in chickens.40 In both birds and mammals, MSCI involves changes in chromatin structure, but the details vary.41 Interestingly, W-inactivation occurs slightly before Z-inactivation in chickens, while Xs and Ys appear to be inactivated simultaneously in mammals.39–41 Moreover, avian MSCI is transitory, while mammalian MSCI is long-lasting with transcription remaining suppressed on both the X and Y throughout the post-meiotic period.39–41 Namekawa and Lee41 speculate that the differences in these post-meiotic effects of MSCI may be due to differences in the mechanisms of dosage compensation in these groups. Regardless, MSCI has independently evolved in both vertebrate XY and ZW taxa, suggesting that it has adaptive function.
Whether MSCI exists in Drosophila has been the source of substantial, periodic controversy over the past four decades.42–45 A recent study shows that while Drosophila species do not have MSCI like that in mammals, genes on the sex chromosomes have drastically reduced expression during spermatogenesis via a different process.45 Further comparative studies of gene expression during gametogenesis are warranted to address the questions: does MSCI always occur when there are sex chromosomes, and how late after the formation of novel sex chromosomes does MSCI evolve?
Dosage compensation and MSCI are fields of intense current study, currently in a state of flux. Despite this uncertainty, we can have confidence that these processes have major effects on the evolution of sex chromosomes, more than what had been previously recognized. Moreover, these processes are inherently epigenetic in nature. That is, they involve heritable changes in gene expression that are not reflected in the DNA sequence. Several other phenomena associated with sex chromosome evolution also involve epigenetic changes; and we will return to the importance of epigenetics at several other places in this review, particularly in our concluding section X.
How do evolutionary processes affect sex chromosomes?
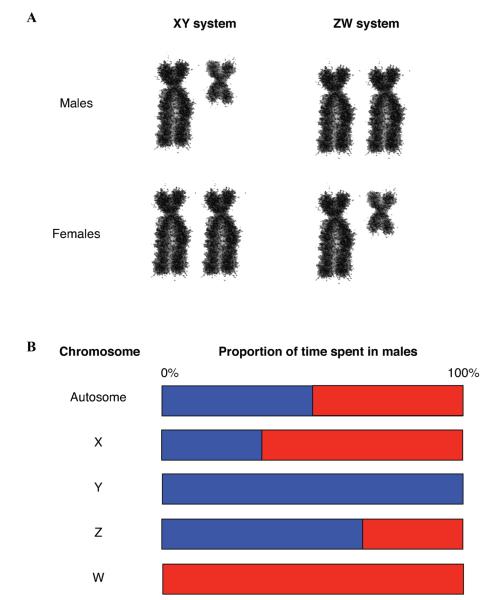
Given the commonly observed 1:1 sex ratio,46 each autosome should be equally represented in both sexes, spending, on average, half the time in males and half in females. In contrast, sex chromosomes will deviate from equal representation. The Y chromosome in XY male heterogametic systems will be present exclusively in males. Likewise, the W chromosome in ZW female heterogametic systems will be present only in females. In XY systems, the X will be present in females two-thirds of the time and in males one-third the time, assuming a 1:1 sex ratio. Finally, the Z chromosome in ZW systems will be present in males two-thirds of the time and in females the other third (see Fig. 1).
Figure 1.
Different types of sex chromosomes. (A) In XY systems males are the heterogametic sex and in ZW systems females are the heterogametic sex. (B) Proportion of time each chromosome spends in males. Male proportions are blue and female proportions are red.
Because sex chromosomes do not spend equal time in each of the sexes, they will experience different effects from evolutionary processes (mutation, random genetic drift, selection, and genomic conflict). Below, we describe the effects these forces have on the substitution rates, the standing genetic variation, and other molecular evolution properties of sex chromosomes and autosomes.47 We summarize these effects in Table 1.
Table 1.
How evolutionary forces affect sex chromosomes and autosomes
| Evolutionary force | Predicted effect | Observations and comments | Key references |
|---|---|---|---|
| Mutation | Mutation rate should be higher in chromosomes that spend more time in males than in females. |
Confirmed in mammals and in birds. Equivocal in flies. |
47 – 56 |
| (Y > Z > A* >W) | Effect strongest in mammals with long generation times. |
||
| Genetic drift | Drift will have larger effect on Y and Z than on A because Y and Z have smaller effective population sizes. |
Generally confirmed. | 47, 57–62 |
| The relative effects of drift on X versus A and Z versus A will depend on reproductive skew (see text). |
|||
| Selection A. SA** selection |
SA variance should be greatest on X and Z. Alleles benefiting females but deleterious to males should be abundant on X. Those benefiting males at expense to females should be less common on X. |
SA selection appears pervasive. Predictions generally confirmed in laboratory experiments with flies. |
63 – 69 |
| B. Dominance and selection |
Negative selection should be more efficient weeding out deleterious X-linked alleles if recessive. Positive selection should also be more efficient fixing beneficial recessive alleles. |
Fly studies confirm that negative selection is more efficient on X. |
47, 70–72 |
| The relative efficiency of positive selection of the X versus A is lineage dependent. |
|||
| C. Additional effects |
Recombination rates may differ between X and A. |
Recombination rate differences will also affect efficiency of positive and negative selection. |
47, 73, 74 |
| Genomic conflict (meiotic drive) |
Drive should be more common on X than A. |
Generally confirmed, but there may be an ascertainment bias. |
16–19, 21, 75–78 |
A stands for autosome.
SA = sexually antagonistic
Mutation
Even at putatively neutral sites, loci on different sex chromosomes evolve at different rates. For instance, autosomal introns have diverged 10.1% between chickens and turkeys (separatedby 28 million years), but their Z-linked and W-linked counterparts have diverged, respectively, 10.9% and 5.7%.48 Between mice and rats, the synonymous substitution rate at X-linked genes is only 81% that of autosomal genes.49 These differences are likely a consequence of two major factors: first, autosomes and sex chromosomes spend different proportions of evolutionary time in males versus females (see above); and second, more germ-line mutations occur in males than in females.50–52
The mutation rate is usually much higher in males than in females because developing male gametes usually undergo many more rounds of replication than do female gametes (reviewed in Ref. 52). In addition, mammalian spermatogenesis is subject to more oxidative stress and less DNA repair per replication than mammalian oogenesis.53 For these reasons, genes that are on chromosomes that are present in males more often than they are in females (i.e., Y and Z) should have a higher mutation rate. Likewise, genes that are on chromosomes that are present in females more often than they are in males (i.e., X and W chromosomes) should have a lower mutation rate. The rank order of mutation rate from highest to lowest is Y > Z > autosomes > X > W.54
Following the formula of Miyata et al.,50 the 19% reduction in synonymous-site substitution rate on the X as compared with the autosomes observed in rodents is consistent with 3.5-fold more mutations in males than in females.49 Consistent with the male-driven mutation hypothesis, this effect is strongest in mammals with longer generation times, and thus, the larger number of replications in the male genome.55
Although male-biased evolution is generally accepted as the major factor influencing differential rates of evolution on sex chromosomes versus autosomes, there are other factors. Autosomes vary in their substitution rate.51 For example, the smaller chromosomes in birds are more gene rich. Based on data from intronic sequences, these “microchromosomes” have higher substitution rates than do the comparable sequences in larger “macrochromosomes.”56 Considerable variation also exists among syntenic blocks within chromosomes.49,51 Complicating matters still further is the conundrum of choosing the appropriate type of sequence to compare (e.g., introns versus silent site changes), an issue that is not fully resolved.51
Random genetic drift
Sex chromosomes and autosomes also experience differential effects from random genetic drift.47 For every mating pair, there are four copies of autosomal genes, three copies of X-linked genes, and one copy of Y-linked genes. If we assume that the sex ratios and effective population sizes of males and females are equal, then in XY systems, the X has an effective size that is 3/4 that of autosomes, while Y chromosomes have effective population sizes that are 1/4 of autosomes. Thus, genetic drift is more powerful and selection less efficient on X-linked genes than on autosomal ones. These effects are even more extreme for Y-linked loci. Similar effects occur for ZW genetic systems. Because effective population size is central to the relative importance of selection versus genetic drift,57,58 rates of adaptive evolution can vary by chromosome.
The above assumes that the variance in reproductive success is the same in males and females (e.g., no reproductive skew exists); but usually, males have a greater variance in reproductive success than females, and thus a lower effective population size. This reproductive skew will thus affect ratios of the effective population sizes of X-linked (or Z-linked) and autosomal genes, bringing the ratio closer to 1 in male heterogametic systems and reducing it below 3/4 in female heterogametic systems.59 Likewise, reproductive skew should cause Y chromosomes to have lower effective population size and different coalescent properties than Z chromosomes.60
Comparisons between X-linked and autosomal variation in humans give a mixed picture. Hammer et al.61 resequenced > 200 kb of noncoding human DNA, finding that the ratio of effective population size for the X chromosome and autosomes was significantly greater than the expected 0.75. By contrast, low-coverage, whole-genome sequence data from the 1000 Genomes Project indicate that X chromosomes are significantly less polymorphic than autosomes after normalizing for mutation rate differences, but vary across populations.62 Furthermore, relative levels of X-linked diversity are reduced near genes.62
Selection
Various forms of selection influence the differential evolution of sex chromosomes and autosomes. One of these is sexual antagonism. Because male and females present “different environments” for natural and sexual selection, sex chromosomes and autosomes can be considered different environments for alleles. Consider sexual antagonism, wherein an allele increases fitness in one sex but reduces it in the other.63–65 An allele that increases male fitness slightly at the expense of greatly diminishing female fitness will be favored on the Y chromosome in a male heterogametic species, but will be selected against it if is on the X chromosome or on the autosomes. An allele that has a positive effect on female fitness, combined with the exactly same negative effect on male fitness, would be neutral on an autosomal locus (assuming a 1:1 sex ratio and large effective population size) but would be favored on the X chromosome. In female heterogametic systems, an allele that increases female fitness at the expense of male fitness would always be favored if it were on the W chromosome, but the relative ratio of the effects on males and females would determine whether it was selectively favored, neutral, or disfavored on the Z or on the autosomes.
Theory also predicts that sexually antagonistic genetic variation will be on the X more often than on the autosomes.63 Rare recessive alleles that have a small beneficial effect in males but a large deleterious effect in females would be selected against if autosomal, but can reach an appreciable equilibrium frequency if X-linked. Similarly, a dominant allele that benefited females greatly, but was somewhat detrimental in males, would go to fixation if autosomal, but could not if X-linked.63,66 Note that this conclusion assumes that the dominance of an allele does not differ between the two sexes.67 Empirical evidence is consistent with theory: X chromosomes are enriched for sexually antagonistic genes in D. melanogaster.68
Evidence of sexually antagonistic genetic variation is also found in natural populations. A wild population of red deer on the Isle of Rum, Scotland, exhibits the signature of segregating X-linked sexually antagonistic genetic variation.69 Here, males with high fitness sire daughters with relatively low fitness, on average, and this negative correlation is likely due to X-linked factors. A recent meta-analysis also showed that sexual antagonistic (SA) selection often acts on wild populations of animals, and can be quite intense.65 Although the pervasiveness of SA selection does not prove that sexually antagonistic variation is also common, SA selection tends to promote the generation and maintenance of such genetic variation. Further studies similar to Ref. 69 investigating whether such X-linked variation exists should be encouraged.
Selection also has different effects on autosomal and sex-linked alleles due to the dominance of those alleles. Because Z and X chromosomes are hemizygous in females and males, respectively, partially or completely recessive alleles on the Z or X would be more exposed to selection when rare as compared with similar autosomal alleles.54,70,71 By contrast, the autosomal alleles are sheltered in heterozygotes when rare. This means that rare recessive, advantageous alleles would be less likely to be lost due to stochastic factors if they are on the Z or X chromosome.71 By the same logic, slightly deleterious and recessive alleles are more efficiently purged when they are on the Z or the X chromosome than when they are on an autosome.
Taking advantage of the 12 sequenced genomes in the genus Drosophila, Singh and colleagues72 compared the relative efficiencies of selection on X-linked and autosomal genes. They found consistent increased efficacy of negative selection for X-linked genes across the phylogeny of Drosophila as revealed by higher codon bias. In contrast, the relative efficacies of positive selection of the X and the autosomes varied; in some lineages, positive selection was more efficient on the X, while in others, it was more efficient on the autosomes.72
Additional factors involving selection and drift
Selection is more efficient and drift is less potent when recombination rates are high.73 Differences in recombination rates will thus affect the relative efficiency of selection at sex-linked and autosomal sites. In organisms like Drosophila where recombination is absent in males, the effective rate of recombination of the X is four-thirds that of autosomes, assuming a 1:1 sex ratio.73 In contrast, in organisms where male and female recombination occurs at equal rates, the X would experience less recombination than the autosomes because it does not have a partner with which to recombine in the heterogametic sex. These changes in recombination will cause small, but measurable, effects on both adaptive74 and background selection.73
The relative strengths of selection and drift also bear on the question: do X-linked DNA sequences evolve faster than autosomal ones? Answers to this question have been mixed, with some studies finding a strong acceleration of X-linked genes relative to autosomal ones, others finding weak or no acceleration, and still others finding X-linked genes evolving slower than autosomal ones.47,59,72 A recent study59 may shed light on why the results varied across studies. They found a strong association between the relative rates of sex chromosome and autosomal evolution, and the ratios of effective populations of the sex chromosomes and the autosomes. In cases where the X (or Z) chromosome had an effective population size well below 0.75 of the autosomes as in birds, the sex chromosome evolved substantially faster than the autosomes. When the X chromosome had an effective population size around 0.75 of the autosomes (weak skew, as in mammals), on average, the X-linked genes evolved only slightly faster than the autosomal ones. In Drosophila, where there is strong reproductive skew and the effective population size of X and autosomes approaches unity, X-linked genes and autosomal ones evolve at comparable rates. These associations are comparable with the fixation of slightly deleterious mutations playing an important role in molecular evolution.57–59
Meiotic drive and genomic conflict
Theory predicts that meiotic drive systems will evolve more readily on sex chromosomes than on autosomes (e.g., see Ref. 25, 26, 75). Empirical evidence seems to support this prediction, although we note the existence of an ascertainment bias to observing meiotic drive on sex chromosomes than on autosomes.25,75 A consequence of sex-linked meiotic drive is a population-wide deviation in the sex ratio away from 1:1, as has been observed in several taxa, including mushroom-feeding species of Drosophila.75 Such a deviation will lead to selection for autosomal suppressors of this drive.25,46 Co-evolution of sex-linked drive and autosomal suppressors could lead to hybrid incompatibility, as predicted by several models.76–78
How do sex chromosomes originate and evolve?
Under the canonical model, sex chromosomes began as a pair of homologous chromosomes that were identical except that one chromosome had a sex-determining locus (SL) that the other lacked3,60,79–82 (see Fig. 2). Sexually antagonistic selection then led to the divergence of the X and Y (Z and W). Alleles that favor the sex determined by the SL at the expense of the other sex are selected to be in linkage disequilibrium with the SL. Chromosomal inversions and any other genetic changes that reduce recombination between the SL and the sexually antagonistic alleles are also favored. Over a considerable period of time, a cluster of tightly linked loci form around the SL on what have become the heteromorphic sex chromosomes.80,81
Figure 2.
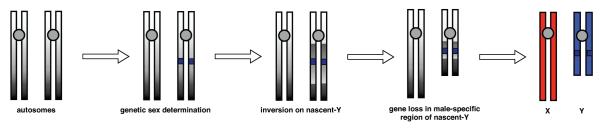
Evolution of sex chromosomes. Under the canonical model, a sex-determining element appears on an autosome. Recombination suppression is favored between SA alleles and the sex-determining factor, leading to the evolution of inversions. Restricted recombination leads to differentiation of sex chromosomes. See text for details.
Lack of recombination between the X and the Y (or the Z and the W) permits genetic degeneration of the Y (W) in the regions that do not recombine.60,80 Like asexual organisms, the non-recombining regions of the Y (W) are subject to Muller’s ratchet, the monotonic progression toward ever more mutations via the stochastic loss of least-mutated chromosomes and continual mutation pressure.80,81 Deleterious mutations can also accumulate via hitchhiking with advantageous alleles, and loss of recombination leads to less efficient purging of the deleterious mutations.80
In addition to an accumulation of deleterious mutations, regions of the genome that do not recombine face reduced potential for adaptive evolution.81 In regions of restricted recombination, advantageous alleles that by chance occur on genetic back-grounds that are loaded with deleterious mutations have a substantially reduced probability of becoming fixed.80,81 In regions of high recombination, by contrast, such an advantageous allele could recombine from the deleterious alleles. This phenomenon, called background trapping, explains why we should expect fewer advantageous alleles to be fixed on the Y.81,83 See Box 1 for more on the degeneration of Y chromosomes.
Box 1. Studies of Y chromosome degeneration.
The human Y chromosome and many other well-studied Y chromosomes are very old (over 100 million years old), and are almost completely degraded. The study of these old chromosomes does not shed light on the early stages and dynamics of Y chromosomal degeneration.
One case of a young Y chromosome is the neo-Y in Drosophila miranda. It is considered a neo-Y chromosome because it form a fusion between the original Y chromosome and an autosome. This fusion occurred approximately one million years ago.165 Despite comparatively young age of the D. miranda neo-Y, this chromosome has already begun to degenerate, both by accumulating repetitive sequence and by the decay of protein-coding genes.92,165
Transposable elements play a large role in the degeneration of the D. miranda neo-Y, with at least one-fifth, and probably closer to one half of the sequence, comprising transposable elements. In contrast, only 1% of its counterpart (the neo-X chromosome) is made up of transposable elements.92 Other new Y chromosomes, such as that in the plant genus Silene, also show an accumulation of transposable elements.166 Such accumulation of transposable elements is expected on Y chromosomes and other nonrecombining regions of the genome; transposable element copy number ordinarily is constrained by natural selection against ectopic exchange, which is much reduced in nonrecombining regions.80 In addition to affecting the expression of genes, transposable elements can also cause chromosomal rearrangements; indeed, one element in the D. miranda neo-Y had caused a gene duplication.165
Protein-coding genes quickly decay after the formation of the neo-Y. About two-fifths of the genes examined in the D. miranda neo-Y are definitively pseudogenes.92,165 The remainder can be translated into a functional protein, and are thus potentially functional. Even these potentially functional genes show signs of possible decay, as they are accumulating nonsynonymous site changes faster than their counterparts on the neo-X. Genes that still maintain a functional copy on the neo-Y evolve more slowly on the X compared with those that have become pseudogenes on the neo-Y, suggesting functional constraint on the X impedes degeneration on the Y.92 There is some evidence for positive selection on this neo-Y, though it is not rampant.165 Theory predicts that adaptive evolution on the Y will be constrained due to Hill-Robertson effects (Refs. 73, 81; see also main text).
In most organisms with highly differentiated chromosomes, a portion of the Y chromosome recombines with the X. This region is known as the pseudoautosomal region (PAR).84 PARs have properties of both autosomes and sex chromosomes, but also are distinct from both types of chromosomes such as having recombination rates substantially higher than autosomes.84 PARs vary in size, depending on how much the X and Y have diverged; two PARs on the human Y make up just under 5% of the total Y chromosome.84 PARs appear to be enriched for traits related to reproductive function in plants of the genus Silene85 as well as in guppies and strawberries.84
In evolutionary comparisons of the X and Y in mammals, genes from certain regions of the Ys all share similar divergences from the X chromosome. These regions, known as “strata” due to the analogy to geological strata,3,60,86 arise as parts of the PAR get subsumed into the nonrecombining section of the Y. These strata likely reflect chromosomal inversions or other rearrangements reducing recombination. Strata have been found in sex chromosomes of birds87 and plants,88 as well as in mammals.86 The nonlinear strata observed in avian Y chromosomes87 may reflect the result of multiple, overlapping inversions. See Box 2 for a discussion of strata in the evolution of primate Y chromosomes.
Box 2. The human Y chromosome.
Human Y chromosomes contain two small PAR that recombine with the X chromosome, and a much larger male-specific region. This male-specific region is divided into the X degenerate region (nonrecombining DNA that was originally derived from the X), which contains the Y-specific genes, and a region consisting of several large palindrome-like repeats that can recombine with themselves.167,168
Within humans, very little variation exists in the protein coding Y-specific genes. In fact, two randomly selected human males will differ by less than a single amino acid sequence on average at the Y-specific genes.169 This is due in part to the lower effective population size of the Y.60 In addition, strong purifying selection has been acting on the Y, as evidenced by the low ratio of nonsynonymous to synonymous site polymorphisms.170
Comparison of the Y-specific portion of the human Y chromosome to the counterparts on the X reveals five strata,168,170 regions of comparable divergence that are most likely the manifestations of recombination-suppressing inversions.86 The youngest of these strata is about 30 million years old, five million years before the split of Old World monkeys and apes.168
A comparison of the rhesus macaque, chimpanzee, and human Y chromosomes reveals long-term stability of the Y.168,170 Since the split with Old World monkeys, Y-specific genes have been lost in the newest strata but not in the older four strata in either the human or rhesus monkey lineage. This is consistent with strong purifying selection operating to maintain function of these genes. Interestingly, along the chimpanzee lineage, several genes in the newest strata have become pseudogenes or have been lost. Sequencing of the bonobo Y chromosome may shed light on when these genes were lost. The functions of the Y-linked genes lost or deactivated in the chimpanzee lineage may have been taken over by new genes elsewhere in the genome.
Noncanonical modes
Not all existing Y chromosomes are derived from the X. New Y chromosomes can be generated by translocations with an autosome attaching to either the X or the Y.89,90 In the process, a neo-X and a neo-Y are formed, with each having the same genetic composition at the time of formation. After formation, neo-Ys are subject to the same forces of degeneration as canonically derived Y chromosomes, while recombination allows the neo-X to not undergo degeneration. The formation of a neo-Y thus allows the study of degeneration at early stages, as has been observed in Drosophila miranda89 (see Box 1). Neo-Y chromosomes are fairly common. In fact, much of the human Y chromosome is technically a neo-Y, as it arose from an autosomal translocation between 80 and 130 million years ago.91,92
Because the few genes that it has do not appear to have originated from X chromosome homologues, the Y chromosome in most Drosophila species may not fit the canonical model.89,90 Moreover, the Drosophila Y has shown a net acquisition of autosomal genes, not a loss of genes as the canonical model predicts.93 For these reasons, Bernardo Carvalho and his colleagues90 argue that the Drosophila Y likely arose from a supernumerary (B) chromosome. The Y chromosomes in tsetse flies and at least two species of Homoptera may also have B chromosome origins.90
Functional effects of gene-poor Ys
Even in organisms that have gene-poor Y chromosomes, the Y still has important effects. For example, Y-linked variation within D. melanogaster affects several phenotypic traits, including position-effect variegation, temperature adaptation, spermatogenesis, and male fitness despite this chromosome’s paucity of genes.94–97 Recently, Lemos, Hartl, and their colleagues have shown myriad effects of this Y chromosome on gene expression.96,98,99 Replacing naturally occurring Y chromosomes within D. melanogaster affected the expression of hundreds to thousands of genes (depending on the statistical cut-off used).96,98 These genes are located on the different chromosomes roughly proportional to the numbers of genes on the chromosomes, with perhaps slightly fewer genes on the X. Genes whose expression were affected by the Y are expressed more in males and less in females than the average gene.98 Which genes are affected (and how) by the Y chromosome also depends on the genetic background, suggesting strong epistatic interactions.98
Much of the effect of the D. melanogaster Y chromosome seems to be due to the ribosomal DNA (rDNA) arrays that are abundant on both the X and the Y. Paredes et al.99 created three Y chromosome lines: two with small amounts of the rDNA arrays deleted (mild deletions) and one (severe deletion) with over half of the arrays removed. The severe deletion may have affected the expression of up to 40% of the genes, with the milder deletions affecting a smaller subset of the genes affected by the severe deletion.99 Genes affected by the Y-linked rDNA found throughout the genome, in a manner in-distinguishable from random and interestingly, not correlated with proximity to heterochromatin. In most of the affected genes, the effects of the deletion were relatively subtle. The gene sensitive to the deletions of rDNA tend to be those that are affected by naturally occurring variation at the Y, and may also have a preferential role in energy metabolism.99
Female heterogametic systems
Birds and snakes are the two major vertebrate groups with ZZ/ZW sex chromosomes (i.e., females are heterogametic). During the 1960s, the Japanese geneticist Susumu Ohno hypothesized that the Z in birds and the X in mammals were homologous. Most studies in the genomic era have not supported Ohno’s contention.1,3,72 Yet some researchers, led by the apparent similarity of some of the platypus X chromosomes to avian Z chromosomes, have called for a reconsideration of the Ohno hypothesis.100 A recent study provides conclusive evidence against the Ohno hypothesis,101 demonstrating that the human X shows no homology to chicken Z. Instead, the human X shows homology to avian autosomes and the avian Z shows homology to a completely different set of mammalian autosomes.101
The chicken Z has half the gene density of chicken autosomes, and the human X has roughly half the gene density of human autosomes.101 Interestingly, the human autosomes that were the precursor of the chicken Z and the chicken autosomes that were the precursor of human X have gene densities that do not differ substantially from other autosomes.101 In both mammals and birds, the gene density of the sex chromosomes decreased due gene loss, and to expansions of repetitive elements. See Box 3 for more on sex determination in birds.
Box 3. How is sex determined in birds?
A single Y-linked gene Sry (sex determining region Y) is crucial in mammalian sex determination; individuals with a functional Sry are males, and individuals without it are females. Is there a comparable gene in birds?
One gene, Doublesex and Mab3-related transcription factor 1 (DMRT1), was proposed as a candidate gene for avian sex determination. As its name suggests, it shows sequence similarity to doublesex in Drosophila and mab3 (male abnormal 3) in C. elegans, both genes that play important roles in sex determination in these animals. DMRT1 also appears to be involved in sex determination in mammals and fish though its role is not exactly known.1,3
Evidence supporting the importance of DMRT1 in avian sex determination came from a study of emus. In these large flightless birds, the Z and W are nearly identical. This is in sharp contrast to most birds wherein the W is much smaller and has many fewer genes than the Z. In emus, DMRT1 is found on the Z, but not on the W.171 This is suggestive, though not conclusive, evidence that DMRT1 is the sex determination gene in birds. One reason that this evidence is not conclusive is that DMRT1 is not the only gene that is on the Z and not the W.172
More recently, Smith et al.173 provided more definitive evidence for DMRT1 being critical to sex determination in birds. They used RNA interference technology to decrease expression of DMRT1 in genetically male chicken embryos. Those embryos that had the most RNA interference had the most feminized gonads. Thus, they determined that DMRT1 expression is needed for development of male gonads. They173 also observed that lower DMRT1 expression (either by being female or by RNA interference) led to a decline or absence of the expression of another gene, SOX9, in the gonads. This finding strongly suggests that SOX9 is a downstream target of DMRT1. The genetically male embryos with altered DMRT1 expression also expressed aromatase, which is usually expressed only in females.173
Female heterogamety is present in several groups of insects, most notably Lepidoptera (butterflies and moths). Lepidopetran female heterogamety is old, as the sister group Trichoptera (caddis flies), which diverged about 190 million years ago, share this mode of sex determination.102 ZW/ZZ predominates in Lepidoptera, although other systems also occur and the sex determination in this group was originally ZZ/ZO, with the W originating from a chromosomal rearrangement.102 In some species like the silkworm moth Bombyx mori, sex is determined by the presence or absence of the W (analogous to the mammalian Y); while other species possess a Z counting mechanism of sex determination, analogous to the X counting in Drosophila sex determination.102 Also analogous to Drosophila, the heterogametic sex in many Lepidopteran species lacks recombination.102,103 While evidence clearly supports multiple origins of male heterogamety in insects,104 it is not yet known whether female heterogamety had multiple origins. Unlike in the case of male heterogametic systems wherein the X chromosome plays a large part in gene movement, there is no evidence of preferential gene movement on and off sex chromosomes in either silkworm moths or birds.104,105
How are genes with sex-biased expression distributed among the chromosomes
In male heterogametic systems, genes that are expressed more in males than in females (male biased) are expected to be on the X chromosome less than expected by chance. This is true in several species of Drosophila.107–110 Genes that are expressed more in females than in males (female biased) are expected to be disproportionately found on the X in Drosophila.107,108
The Drosophila X does not just have fewer than expected male-biased genes; it also is particularly depauperate for genes that are the most highly expressed in testes.110 One simple explanation for this pattern is that the genes that are highly expressed in the testes are under greater selective pressure to move to the autosomes. A comparison of expression patterns across closely related species, however, reveals that changes in gene expression rather than gene movement is primarily responsible for autosomal genes that have recently evolved male-biased gene expression.110
An alternative explanation involves dosage compensation by hyper transcription of the X chromosome in XY males (or the Z in ZW females). If an upper limit to transcription exists, then genes that are highly expressed in males would be more likely to be near this limit when they are on an X chromosome than when they are autosomal. If this is so, then there would be selection against highly expressed X-linked genes, and thus the paucity of male-expressed genes should be more severe for highly expressed genes than those with lower expression. Comparative studies with other taxa would shed light on the generality of this pattern, and could determine whether it is something explainable as a consequence of Drosophila-like dosage compensation. For instance, this explanation is not likely in mammals.110
A recent study suggests another mechanistic role by which dosage compensation can affect patterns of gene expression on the Drosophila X chromosome.34 Recall that in Drosophila dosage compensation, binding of the MSL complex to high affinity sites (HAS) on the X gene elevates expression on the male X. This study showed that genes expressed more in males were much less likely to be located near an HAS than in other regions of the X.34 In contrast, genes expressed more in females or equally between the sexes did not show a decreased density near an HAS. The avoidance of HAS by male-biased genes contributes to the relative lack of male-expressed genes on the X.34 Although, as we will see, other factors also contribute to the lack of X-linked male-biased genes, this study does show that dosage compensation affects the nature of genes found on the X. It also raises the prospect that hybrid incompatibility could arise from disruptions in dosage compensation.
The demasculanization of the X appears to evolve quickly. The neo-X chromosome of D. pseudoobscura, which evolved about ten million years ago, already shows a substantial deficit of male-expressed genes.109 The loss of male-expressed genes on the neo-X is mostly due to the extinction of male-biased genes on the X (and birth of new male-expressed genes on the autosomes, as well as movement of the male-expressed genes from the neo-X to the autosomes.109 Changes in the sex-bias expression of individual genes had little to do with the demasculanization.109
Similar patterns are observed in other male heterogametic insects. In the mosquito, Anopheles gambiae, which has an X that is homologous to the one in Drosophila104 testes-expressed genes are at a lower than expected frequency on the X.111 The X chromosome of the flour beetle Tribolium castaneum arose independently from the one in flies and mosquitoes.104 It has fewer male-biased and more female-biased genes than expected.112
Studies in mammals suggest that MSCI plays an important part in the evolution of patterns of genes that are expressed on the X.113–115 Genes in mice that are expressed early in spermatogenesis are overly abundant on the X, but ones that are expressed later are found on the X much less frequently than expected,113 consistent with demasculanization of the X being a consequence of MSCI114 (but see Ref. 109). Another study115 found that mammalian X chromosomes have undergone two bursts of gene gain: an early burst predated the divergence of eutherian mammals and marsupials, and a later burst occurred after the human/chimp split and the mouse/rat split. In mammals, the expression patterns of young X-linked genes were found to be male-biased, in contrast to old X-linked genes, which were not male-biased.115 Furthermore, young X-linked genes in mammals appear to be unaffected by MSCI, while old X-linked genes are often silent in meiosis.
Less is known about the chromosomal patterns of sex-specific genes in taxa with female heterogametic systems. One study found that the Z chromosome of silkworm moths had a greater than expected number of testes-specific genes.116 This is the expected converse of the demasculanization of X chromosomes in taxa with male heterogamety. It is perhaps noteworthy that even though the same pattern of sex-biased gene distribution exists in some ZW systems there are no ZW systems with “global” Z dosage compensation so far.3
What roles do sex chromosomes play in hybrid incompatibility?
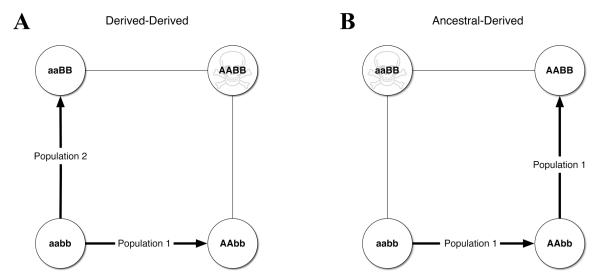
Before discussing how the implications of sex chromosome evolution for hybrid incompatibility, we briefly describe the general framework for how hybrid incompatibility usually evolves. During the first half of the twentieth century, Bateson,117 Dobzhansky,118 and Muller119 recognized that hybrid incompatibility generally requires at least two genetic changes. Thus, according to the Bateson–Dobzhansky–Muller (BDM) model, hybrid incompatibility is a failure of the proper interaction of genes (see Fig. 3).
Figure 3.
Two types of BDM incompatibilities. BDM incompatibilities involve epistatic interactions between alleles at two different loci. Alleles at locus 1 are a and A, and alleles at locus 2 are b and B. Recessive autosomal BDM incompatibilities are depicted. However, these incompatibilities can also be dominant and/or sex-linked. The ancestral genotype is aabb. (A) Derived-derived incompatibilities. Each of two divergent populations fixes an incompatible allele at a different locus. Upon secondary contact these alleles interact, causing fitness to be reduced. (B) Ancestral-derived incompatibilities. A single population undergoes sequential fixation events at two different loci. The intermediate genotype (AAbb) does not have reduced fitness. Upon secondary contact incompatible ancestral (a) and derived (B) alleles interact, causing fitness to be reduced.
Starting in the late 1990s, evolutionary geneticists have characterized a growing list of genes that contribute to hybrid incompatibility. Several recent reviews have discussed these genes and their evolution.20–23 Although more progress has been made in identifying individual genes involved in hybrid incompatibility than interacting sets of genes, the BDM model appears to be holding up well (see Ref. 20; but also see Ref. 120). Because more than a single genetic change is needed to yield BDM incompatibilities, theory predicts that the number of genes involved in hybrid incompatibility should accumulate faster than linear with genetic divergence, a phenomenon known as the “snowball.”121 Recent data from both tomatoes122 and Drosophila123 support the existence of such a snowball.
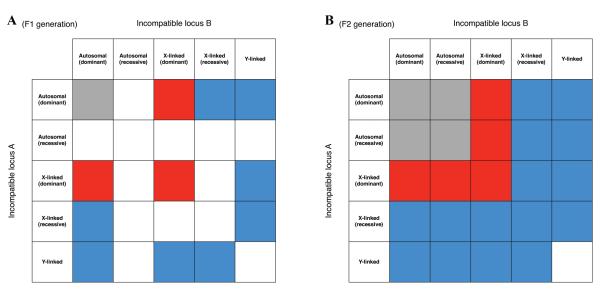
Importantly, BDM incompatibilities can involve sex-linked alleles. When incompatibilities involve sex-linked alleles, sex-biased patterns of sterility and/or lethality arise (see Fig. 4). These sex-biased patterns depend on whether incompatibilities are dominant or recessive. Dominant incompatibilities result in an excess mortality and/or sterility for the homogametic sex, and recessive incompatibilities result in reduced fitness of the heterogametic sex (i.e., Haldane’s rule-like patterns arise, see below and Fig. 4). X-autosome interactions can also result in hybrids of reciprocal crosses differing in fertility or viability, a pattern sometimes called Darwin’s corollary to Haldane’s rule.19
Figure 4.
BDM incompatibilities and sex-linked alleles. Incompatibilities are shown for the F1 generation (panel A) and for the F2 generation (panel B). An XY system is assumed, and colors reveal whether hybrid compatibilities are sex biased. Gray: both sexes are affected equally. White: neither sex affected. Red: greater fitness reduction in the homogametic sex (females). Blue: greater fitness reduction in the heterogametic sex (males), consistent with Haldane’s rule. Sex-biased patterns in the F1 generation exhibit parent-of-origin effects. Individuals that are homozygous for recessive alleles can be observed in the F2 generation.
Recent theoretical work also indicates that standing genetic variation can contain X-autosome incompatibilities.124 This possibility arises because incompatible X and autosomal alleles are able to segregate at relatively high frequencies (on the order of for recessive incompatibilities, where μ is the mutation rate and s is the selection coefficient). Frequencies of incompatible alleles can vary across populations. For example, one population can have a high frequency X-linked allele that is incompatible with low frequency autosomal alleles. Meanwhile, a second population can have an incompatible autosomal allele at high frequency and an incompatible X-allele at low frequency.
Secondary contact between such divergent populations can result in a form of interpopulation hybrid incompatibility, and due to dominance/recessivity of X-linked alleles, this incompatibility can be sex-biased.124 Because the effects of recessive X-autosome incompatibilities are masked in heterozygous females, Haldane’s rule-like patterns can occur during the early stages of speciation. Theory also indicates that X-autosome epistasis facilitates reinforcement when fitness hits occur in males, but not when they occur in females.125
Many empirical examples of negative epistasis involving sex-linked alleles exist. For instance, crosses between wild-derived strains of house mice, Mus musculus and Mus domesticus, result in males with reduced fitness.126 Because the extent of male sterility in these hybrid mice depends on which parent supplies an X chromosome, interactions involving X chromosomes have been implicated in the early stages of Haldane’s rule. In Drosophila, hybrid inviability arises from interactions between the autosomal nucleoporin gene Nup160 of D. simulans and X-linked genes of D. melanogaster.127 Other X–A interactions that underlie hybrid incompatibility in Drosophila have been mapped.4,128 Note also that X linked factors can interact in X–X interactions.4,129 Deleterious epistatic fitness effects can also involve variation within species. Using D. melanogaster, a recent study revealed that flies with X chromosomes and autosomes from different geographic locations exhibit large levels of synthetic sterility.130 Similarly, X-autosome incompatibilities underlie Haldane’s rule-like patterns in a possible case of incipient speciation involving divergent populations of the flour beetle Tribolium castaneum.131
What explains Haldane’s rule?
The explanation for Haldane’s6 rule, wherein the hemizygous sex is more adversely affected in F1 hybrid crosses, has been the subject of considerable attention and debate. Several explanations have been presented, none of which fully explains the rule by itself.4,8 Most of these explanations are the consequence of the properties of sex chromosomes.
The dominance model
A popular explanation for Haldane’s rule is the dominance model.132–134 This model, which is an extension of ideas that Muller proposed, posits that the alleles that contribute to hybrid incompatibility are at least partially recessive in hybrids. Thus, if these alleles are X-linked, their deleterious consequences will affect hemizygous hybrid males more than they will affect heterozygous hybrid females. Based on the same logic, Z-linked recessive alleles will be more deleterious in hemizygous females than in heterozygous males in hybrids in systems with female heterogamety. Consistent with the expectations of the dominance model, species pairs of Drosophila with large X chromosomes evolve Haldane’s rule more rapidly than those with smaller X chromosomes.135 Also consistent with this dominance model is the observation that haplodiploid species, which lack sex chromosomes, exhibit a form of Haldane’s rule wherein hybrid haploid males are affected more than their diploid sisters.12,136
Although the dominance model explains a substantial part of the observations surrounding Haldane’s rule and associated phenomena, it does not explain all. Particularly striking is the fact that exceptions to Haldane’s rule are rather common for viability aspect in flies and in mammals (wherein F1 males are viable, but F1 females are inviable), but are rare for the sterility aspect.4,8 If the dominance model were the only major explanation for Haldane’s rule, exceptions to the rule would be rare for both the sterility and the inviability aspects. One plausible explanation for these exceptions is that they arise from interactions between the X chromosome of one species and maternal effects from the other. This appears to be the case in Drosophila,137 but there is little evidence for this process outside this genus.
Recent evidence that marsupials exhibit Haldane’s rule also challenges the generality of the dominance model: because marsupials perform dosage compensation by inactivating the paternal X chromosome, the female F1 hybrids as unbalanced as their brothers.30 Thus, the dominance model alone cannot fully explain Haldane’s rule. Three other models have been postulated: “faster male,” “faster X,” and “faster heterogametic sex/genomic conflict.” We review these below.
The “faster male” model
In both Drosophila and mammals, there are more total cases of the sterility aspect of Haldane’s rule than for the inviability aspect.4,8,138 Comparative studies also show that hybrid sterility also evolves faster than hybrid inviability in Drosophila8,138,139 and in frogs.141 Moreover, many more genetic regions contribute to hybrid male sterility than hybrid female sterility or hybrid inviability,8,9,14,16 despite viability being a larger mutational target within species.8 These observations lay the foundation of the “faster male” model, which states that male reproductive systems evolve faster than female reproductive systems and most somatic tissues. Therefore, the incompatibility in hybrids should manifest mainly as male sterility.8 The faster male model can only explain Haldane’s rule in XY systems; operation of the faster male model in ZW systems would run counter to the expectations of Haldane’s rule.134
The faster male model could arise from sexual selection driving the divergence that causes sterility in hybrids.8,9 Sexual selection is a powerful force, and the variance in male reproductive success is usually greater than the variance in female reproductive success. Thus, the opportunity for sexual selection is greater in males than in females.141 Consistent with this explanation, genes that are expressed in male reproductive systems are among the fastest evolving genes in both mammals142 and flies.143
Particular properties of spermatogenesis in male heterogametic taxa could also accelerate the evolution of hybrid male sterility.8 Although this hypothesis has less direct support, the growing importance of the proper inactivation of the sex chromosomes during meiosis, as well as dosage compensation, suggests that we should not dismiss this hypothesis out of hand.
The “faster X” model
Another possible explanation for Haldane’s rule is “the faster X” model, which is defined as genetic changes accumulating at a faster rate on the X chromosome than on the autosomes.4,71 This faster X explanation is distinct from the “large X” effect, which is the more rapid accumulation of hybrid incompatibility factors on the X as compared with the autosomes. The large X effect, which is strongly supported by genetic data in Drosophila,16 could be explained by the faster X model, but it need not be.4,71
Recent studies (Ref. 59; see above) show that the X usually evolves somewhat faster than autosomes at the DNA sequence level, though the extent of this effect varies across taxa and even among different studies of the same taxa. Taken at face value, these data would support a role for the faster X contributing to Haldane’s rule and the large X effect in speciation. However, the faster X at the sequence level is strongest when the effective population size of the sex chromosome is smallest relative to autosomes, suggesting that that the faster X is the result of the accumulation of slightly deleterious substitutions.59 In contrast, many of the genes involved in hybrid incompatibility show the hallmark of being driven by positive selection, whether natural selection or genomic conflict.20–23 As of this writing, the faster X model has little empirical support.
Faster heterogametic sex/ genomic conflict
If genomic conflict over the sex ratio is common and hybrid incompatibility frequently arises as a consequence of such conflict, then we would expect hybrid incompatibility to affect the heterogametic sex in taxa with both XY and ZW sex determining systems.15 Consistent with theory, sex-ratio distorters appear to be more common than autosomal distorters, though an ascertainment bias exists (Ref. 75; see above). Support for this model comes from the Overdrive gene, which causes both sterility and sex-ratio distortion in hybrids of Drosophila pseudoobscura.144 Moreover, most of the genes that interact with Overdrive also contribute to both the sterility and the drive.145 Other more indirect studies also appear to support the contention that genomic conflict may accelerate the evolution of hybrid incompatibility in the heterogametic sex.20–23 However, more research is needed to determine the relative importance of this factor.
Summary
Three of the four models (dominance, faster X, faster heterogametic sex) explaining Haldane’s rule derive directly from properties of sex chromosomes. Even the faster male model could arise indirectly from processes stemming from the hemizygosity of sex chromosomes if it is the result of special properties of spermatogenesis in male heterogametic systems.
What are the effects of the sex chromosomes on gene expression in hybrids?
Recall that fewer male-expressed genes are located on the X chromosome in some groups, including Drosophila. How does demasculinization of the X relate to speciation? Given that hemizygous male sterility genes are less likely to be mutational targets in a demasculinized X, this might slow down the accumulation of hybrid male sterility factors and retard Haldane’s rule.
Several studies, especially in Drosophila and mammals, have also examined gene expression in hybrids. One of the hopes of these studies was that they could reveal genes that contribute to hybrid incompatibility.146 We caution that it is often difficult to disentangle cause and consequence in the gene expression–hybrid incompatibility relationship. Genes that contribute to hybrid incompatibility may cause deleterious effects in hybrids because they are expressed inappropriately. It is also possible that some genes, which do not have a causal effect on hybrid incompatibility, may be misexpressed due to the breakdown of cellular processes associated with sterility or inviability (Ref. 146; see below).
In crosses between Mus musculus and M. domesticus, the fertility of F1 hybrids depends on both the direction of cross and the parental genotypes.126 A recent study by Good and colleagues147 took advantage of the differences in fertility among different types of hybrids to link hybrid fertility to gene expression. They considered the 902 genes that differed significantly between the reciprocal hybrids and at least one of the parental lines. In comparison to the fertile hybrids, sterile hybrids showed a pattern of overexpression of a disproportionately large number of X-linked genes. Moreover, sterility-correlated genes showed an overrepresentation of postmeiotic genes, but fewer than expected meiotic genes. Good et al.147 (p. 8) conclude, “these data support the hypothesis that spermatogenic gene regulation on the X chromosome is particularly sensitive to incompatible interactions between the divergent genomes of M. musculus and M. domesticus.” They also identify two candidate genes that lie near the top of the cascade that goes awry in hybrids: (1) DNA methyltransferase 3A, an essential spermatogenesis gene, which may play a role in MSCI or later silencing of the X in spermatogenesis; and (2) the transcription factor Brwd1 that is involved in chromatin remodeling during spermatogenesis. Interestingly, a lower proportion of X-linked genes than autosomal ones changed expression patterns between species. This conservation of X-linked gene expression is in sharp contrast to the other finding that protein evolution (as measured by the ratio of nonsynonymous to synonymous changes) is evolving faster on X-linked testes-expressed genes than on autosomal genes.147 Similarly, studies in Drosophila find that the X has a greater than expected number of genes that are overexpressed in hybrids, but fewer that are underexpressed.148,149
What effect does X-linked genes that contribute to hybrid male sterility have on gene expression in hybrids? The X-linked Odysseus (Ods) is a rapidly evolving gene that contributes to the male sterility seen in hybrids between D. mauritiana and D. simulans.150 Introgression of Ods also causes a disproportionately large number of autosomal genes to alter their expression, confirming the importance of X-autosomal interactions.151 Interestingly, Ods also interacts with heterochromatin on the Y chromosome in sterile hybrids.152 The nature of any of these interactions, and whether they are cause or consequence of hybrid sterility, is not known.
Introgressions of heterospecific Y chromosomes also affect gene expression in hybrids. When intro-gressed into the genetic background of D. simulans, the D. sechellia Y has little or no effect on viability, but reduces male offspring production and sperm competiveness.153,154 A recent study shows that the Y introgression also decreases that expression of a number of genes, including many spermatogenesis genes.154 Interestingly, the introgression also up-regulates genes involved with immune function, suggesting a tradeoff between immune function and spermatogenesis.154 In light of the myriad epigenetic effects of intraspecific Y chromosome replacements,94–99 such effects in hybrids should not be surprising.
Do sex chromosomes promote hybrid incompatibility?
Several authors (see, e.g., Refs. 20, 26) have suggested that due to the associated genomic conflict with sex chromosomes, the presence of heteromorphic sex chromosomes may promote the evolution of hybrid incompatibility, and hence, speciation. What, if any, evidence supports this thesis?
An ideal test of this idea would include having a metric closely tied to hybrid incompatibility, such as the index of postzygotic isolation developed by Coyne and Orr,139 as well as taxa that have had several independent evolutions of heteromorphic sex chromosomes or losses of such chromosomes. Currently, we lack these conditions.
Two suggestive pieces of evidence, however, support this model. First haplodiploid species, which do not have specialized sex chromosomes, appear to evolve hybrid incompatibility more slowly than do other insects, although they exhibit a variant of Haldane’s rule (see above).136 Second, sex chromosomes are common in squamates (lizards and snakes), but rare in turtles and crocodiles. Consistent with the model, squamates have diversified more rapidly than turtles and crocodiles.155,156 Diversification rates are affected by extinction as well as forms of speciation unrelated to hybrid incompatibility. Furthermore, the number of independent transitions in sex chromosome status is meager. More data both on rates of accumulation of hybrid incompatibility and sex determination systems are needed to more definitely address this question. If we can confirm that the presence of heteromorphic sex chromosomes accelerates speciation via increasing the rate at which hybrid incompatibility evolves, species selection would be acting on the species-level trait heteromorphic sex chromosomes.4
What are the connections between sex chromosomes, hybrid incompatibility, and epigenetics?
The specter of epigenetics looms large in many of the recent studies of hybrid incompatibility, especially those involving sex chromosomes. Following Waddington’s view,157,158 Goldberg et al.159 (p. 635) state: “Epigenetics, in a broad sense, is a bridge between genotype and phenotype—a phenomenon that changes the final outcome of a locus or chromosome without changing the underlying DNA sequence.”
Earlier in this review, we noted several instances of such broad-sense epigenetics being involved in sex chromosome evolution and possibly hybrid incompatibility. Despite having few coding genes, the Drosophila Y chromosome affects both gene expression patterns and phenotypic traits in pure species95–99 and in hybrids between species,154 and apparently does so through epigenetic mechanisms. Several genes that are known to contribute to hybrid incompatibility are heterochromatic or interact with heterochromatin.20–23 Thus, chromatin structure may play a larger than previously expected role in hybrid incompatibility. Moreover, despite their profound differences, both mammalian and Drosophila dosage compensation involve similar forms of epigenetic mechanisms.27,160 Likewise, MSCI and related silencing of unpaired chromosomes during meiosis also involve epigenetic mechanisms.38 Emerging evidence suggests that the breakdown of dosage compensation34 and MSCI147 may be important contributors to the sterility and inviability seen in hybrids. If so, then epigenetics and hybrid incompatibility are even more tightly intertwined.
We are far from the first to link epigenetics to hybrid incompatibility. During the past 30 years, several authors have postulated epigenetic mechanisms for the evolution of hybrid sterility and inviability (e.g., Refs. 161 and 162). These epigenetic arguments have largely fallen on deaf ears in the main-stream evolution community. Indeed, the leading book on speciation4 does not even have an entry on “epigenetics” or on “chromatin.” We contend that although the specific mechanisms postulated by the past champions of epigenetics may be incorrect or limited in scope, they may have been correct about the larger picture. These arguments for epigenetic mechanisms of speciation deserve a close second examination.
Acknowledgments
Two anonymous reviewers and the editors provided many useful suggestions, for which we are grateful. We thank Jeff Demuth and Daven Presgraves for graciously supplying us with prepublication manuscripts and for insightful comments on an early draft of this manuscript. We also thank Wilfred Haerty for supplying supplemental materials from his publication. JL is supported by an NIH NRSA postdoctoral fellowship (F32HG006648–01).
Footnotes
Conflicts of interest The authors declare no conflicts of interest.
References
- 1.Charlesworth D, Mank JE. The birds and the bees and the flowers and the trees: lessons from genetic mapping of sex determination in plants and animals. Genetics. 2010;186:9–31. doi: 10.1534/genetics.110.117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graves JAM, Peichel CL. Are homologies in vertebrate sex determination due to shared ancestry or to limited options? Genome Biol. 2010;11:205–216. doi: 10.1186/gb-2010-11-4-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellegren H. Sex-chromosome evolution: recent progress and the influence of male and female heterogamety. Nat. Rev. Genet. 2011;12:257–266. doi: 10.1038/nrg2948. [DOI] [PubMed] [Google Scholar]
- 4.Coyne JA, Orr HA. Speciation. Sinauer Associates, Inc.; Sunderland, MA: 2004. [Google Scholar]
- 5.Presgraves DC. Sex chromosomes and speciation in Drosophila. Trends Genet. 2008;24:336–343. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haldane JBS. Sex ratio and unisexual sterility in animal hybrids. J. Genet. 1922;12:101–109. [Google Scholar]
- 7.Coyne JA, Orr HA. Two rules of speciation. In: Otte D, Endler JA, editors. Speciation and Its Consequences. Sinauer Associates, Inc.; Sunderland, MA: 1989. pp. 180–207. [Google Scholar]
- 8.Wu CI, Davis AW. Evolution of postmating reproductive isolation: the composite nature of Haldane’s rule and its genetic bases. Am. Naturalist. 1993;142:187–212. doi: 10.1086/285534. [DOI] [PubMed] [Google Scholar]
- 9.Wu CI, Johnson NA, Palopoli MF. Haldane’s rule and its legacy: why are there so many sterile males? Trends Ecol. Evol. 1996;11:281–284. doi: 10.1016/0169-5347(96)10033-1. [DOI] [PubMed] [Google Scholar]
- 10.Laurie CC. The weaker sex is heterogametic: 75 years of Haldane’s rule. Genetics. 1997;147:937–951. doi: 10.1093/genetics/147.3.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orr HA. Haldane’s rule. Annu. Rev. Ecol. Syst. 1997;28:195–211. [Google Scholar]
- 12.Schilthuizen M, Giesbers MCWG, Beukeboom LW. Haldane’s rule in the 21st century. Heredity. 2011;107:95–102. doi: 10.1038/hdy.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brothers AN, Delph LF. The generality of Haldane’s rule is to plants with sex chromosomes. Evolution. 2010;64:3643–3648. doi: 10.1111/j.1558-5646.2010.01095.x. [DOI] [PubMed] [Google Scholar]
- 14.True JR, Weir BS, Laurie CC. A genome-wide survey of hybrid incompatibility factors by the introgression of marked segments of Drosophila mauritiana chromosomes into Drosophila simulans. Genetics. 1996;142:819–837. doi: 10.1093/genetics/142.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tao Y, Hartl DL. Genetic dissection of hybrid incompatibilities between Drosophila simulans and Drosophila mauritiana. III. Heterogenous accumulation of hybrid incompatibilities, degree of dominance, and implications for Haldane’s rule. Evolution. 2003;57:2580–2598. doi: 10.1111/j.0014-3820.2003.tb01501.x. [DOI] [PubMed] [Google Scholar]
- 16.Masly JP, Presgraves DC. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biology. 2007;5:1890–1898. doi: 10.1371/journal.pbio.0050243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sperling FAH. Sex-linked genes and species differences in Lepidoptera. Can. Entomologist. 1994;126:807–818. [Google Scholar]
- 18.Prowell DP. Sex linkage and speciation in Lepidoptera. In: Howard DJ, Berlocher SH, editors. Endless Forms: Species and Speciation. Oxford University Press; New York: 1998. pp. 309–319. [Google Scholar]
- 19.Turelli M, Moyle LC. Asymmetric postmating isolation: Darwin’s corollary to Haldane’s rule. Genetics. 2007;176:1059–1088. doi: 10.1534/genetics.106.065979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson NA. Hybrid incompatibility genes: remnants of a genomic battlefield? Trends Genet. 2010;26:317–325. doi: 10.1016/j.tig.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Presgraves DC. The molecular genetic basis of species formation. Nat. Rev. Genet. 2010;11:175–180. doi: 10.1038/nrg2718. [DOI] [PubMed] [Google Scholar]
- 22.Maheshwari S, Barbash DA. The genetics of hybrid incompatibilities. Ann. Rev. Genet. 2011;45:331–355. doi: 10.1146/annurev-genet-110410-132514. [DOI] [PubMed] [Google Scholar]
- 23.Sawamura K. Chromatin evolution and molecular drive in speciation. Int. J. Evol. Biol. 2012 doi: 10.1155/2012/301894. doi:10.1155/2012/301894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kocher TD. Adaptive evolution and explosive speciation. Nat. Rev. Genet. 2004;5:288–298. doi: 10.1038/nrg1316. [DOI] [PubMed] [Google Scholar]
- 25.Burt A, Trivers R. Genes in Conflict: The Biology of Selfish Genetic Elements. Belknap Press; Cambridge, MA: 2006. [Google Scholar]
- 26.Meiklejohn CD, Tao Y. Genetic conflict and sex chromosome evolution. Trends Ecol. Evol. 2010;25:215–223. doi: 10.1016/j.tree.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marin I, Siegal ML, Baker BS. The evolution of dosage compensation. Bioessays. 2000;22:1106–1114. doi: 10.1002/1521-1878(200012)22:12<1106::AID-BIES8>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 28.Lyon M. Gene action in the X chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 29.Cooper D, Vandeberg J, Sharman G, Poole W. Phosphoglycerate kinase polymorphism provides further evidence for paternal X inactivation. Nature. 1971;230:155–157. doi: 10.1038/newbio230155a0. [DOI] [PubMed] [Google Scholar]
- 30.Watson E, Demuth J. Haldane’s rule in marsupials: what happens when both sexes are functionally hemizygous? J. Heredity. 2012 doi: 10.1093/jhered/esr154. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller HJ. Further studies on the nature of causes of gene mutations. Proc. 6th Int. Congr. Genet. 1932;1:213–235. [Google Scholar]
- 32.Baker BS, Marin I, Gorman M. Dosage compensation in Drosophila. Ann. Rev. Genet. 1994;28:491–521. doi: 10.1146/annurev.ge.28.120194.002423. [DOI] [PubMed] [Google Scholar]
- 33.Smith ER, Allis CD, Lucchesi JC. Linking global histone acetylation to the transcription enhancement of XChromosomal genes in Drosophila males. J Biol. Chem. 2001;276:31483–31486. doi: 10.1074/jbc.C100351200. [DOI] [PubMed] [Google Scholar]
- 34.Bachtrog D, Toda NRT, Lockton S. Dosage compensation and demasculinization in Drosophila. Curr. Biol. 2010;20:1476–1481. doi: 10.1016/j.cub.2010.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mank JE, Hosken DJ, Weddell N. Some inconvenient truths about sex chromosome dosage compensation and the potential role of sexual conflict. Evolution. 2011;65:2133–2144. doi: 10.1111/j.1558-5646.2011.01316.x. [DOI] [PubMed] [Google Scholar]
- 36.Xiong YY, Chen XS, Chen ZD, et al. RNA sequencing shows no dosage compensation of the active X chromosome. Nat. Genet. 2010;42:1043–1047. doi: 10.1038/ng.711. [DOI] [PubMed] [Google Scholar]
- 37.Deng, et al. 2011.
- 38.Turner JMA. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- 39.Namekawa SH, VandeBerg JL, McCarrey JR, Lee JT. Sex chromosome silencing in the marsupial male germ line. Proc. Nat. Acad. Sci. 2007;104:9730–9735. doi: 10.1073/pnas.0700323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schoenmakers S, Wssenaar E, Hoogerbrugge JW, et al. Female meiotic sex chromosome inactivation in chicken. PLoS Genet. 2009;5:e1000466. doi: 10.1371/journal.pgen.1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Namekawa SH, Lee JT. XY and ZW: is meiotic sex chromosome evolution inactivation the rule in evolution. PLoS Genet. 2009;5:e1000493. doi: 10.1371/journal.pgen.1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lifschytz E. X-chromosome inactivation: an essential feature of normal spermiogenesis in male heterogametic organisms. In: Beatty RA, Gluecksohn-Waelsch S, editors. Proceedings of the International Symposium on the Genetics of the Spermatozoon. Bogtrykkeriet Forum; Copenhagen: 1972. pp. 223–232. [Google Scholar]
- 43.Lifschytz E, Lindsley DL. The role of X-chromosome inactivation during spermatogenesis. Proc. Natl. Acad. Sci. 1972;69:182–186. doi: 10.1073/pnas.69.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hense W, Baines JF, Parsch J. X chromosome inactivation during Drosophila spermatogenesis. PLoS Biol. 2007;5:2288–2295. doi: 10.1371/journal.pbio.0050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meiklejohn CD, Landeen EL, Cook JM, et al. Sex chromosome-specific regulation in the Drosophila male germline but little evidence for chromosomal dosage compensation or meiotic inactivation. PLoS Biol. 2011;9:e1001126. doi: 10.1371/journal.pbio.1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fisher RA. The Genetical Theory of Natural Selection. Clarendon Press; Oxford: 1930. [Google Scholar]
- 47.Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nat. Rev. Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- 48.Axelsson E, Smith NGC, Sundstrom H, et al. Male-biased mutation rate and divergence in autosomal, Z-linked, and W-linked introns of chicken and turkey. Mol. Biol. Evol. 2004;21:1538–1247. doi: 10.1093/molbev/msh157. [DOI] [PubMed] [Google Scholar]
- 49.Malcom CM, Wyckoff GJ, Lahn BT. Genic mutation rates in mammals: local similarity, chromosomal heterogeneity, and X-versus-autosome disparity. Mol. Biol. Evol. 2003;20:1633–1641. doi: 10.1093/molbev/msg178. [DOI] [PubMed] [Google Scholar]
- 50.Miyata T, Hayashida H, Kuma K, et al. Male-driven molecular evolution: a model of nucleotide sequence analysis. Cold Spring Harb. Symp. Quant. Biol. 1987;52:863–867. doi: 10.1101/sqb.1987.052.01.094. [DOI] [PubMed] [Google Scholar]
- 51.Ellegren H. Characteristics, causes, and evolutionary consequences of male-biased mutation. Proc. Roy. Soc. Lond. B. 2007;274:1–10. doi: 10.1098/rspb.2006.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson Sayres MA, Makova KD. Genome analysis substantiate male mutation bias in many species. Bioessays. 2011;33:938–945. doi: 10.1002/bies.201100091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aitken RJ, Graves JAM. Human spermatozoa: the future of sex. Nature. 2002;415:963. doi: 10.1038/415963a. [DOI] [PubMed] [Google Scholar]
- 54.Kirkpatrick MK, Hall DW. Male-biased mutation, sex linkage, and the rate of adaptive evolution. Evolution. 2004;58:437–440. [PubMed] [Google Scholar]
- 55.Wilson Sayres MA, Venditti C, Pagel M, Makov KD. Do variations in substitution rates and male mutation bias correlate with life-history traits? A study of 32 mammalian genomes. Evolution. 2011;65:2800–2815. doi: 10.1111/j.1558-5646.2011.01337.x. [DOI] [PubMed] [Google Scholar]
- 56.Axelsson E, Webster MT, Smith NGC, et al. Comparison of the chicken and turkey genomes reveals a higher rate of nucleotide divergenceon microchromosomes than macrochromosomes. Genome Res. 2005;15:120–125. doi: 10.1101/gr.3021305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohta T, Gilespie JH. Development of neutral and nearly neutral theories. Theor. Popul. Biol. 1996;49:128–142. doi: 10.1006/tpbi.1996.0007. [DOI] [PubMed] [Google Scholar]
- 58.Nei M, Suzuki Y, Nozawa M. The neutral theory of molecular evolution in the molecular era. Ann. Rev. Genom. Human Genet. 2010;11:265–289. doi: 10.1146/annurev-genom-082908-150129. [DOI] [PubMed] [Google Scholar]
- 59.Mank JE, Vicoso B, Berlin S, Charlesworth B. Effective population size and the faster-X effect: empirical results and their interpretation. Evolution. 2009;64:663–674. doi: 10.1111/j.1558-5646.2009.00853.x. [DOI] [PubMed] [Google Scholar]
- 60.Mank JE. Small, but mighty: the evolutionary dynamics of W and Y sex chromosomes. Chromosome Res. 2012;20:21–33. doi: 10.1007/s10577-011-9251-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hammer MF, Mendez FL, Cox MP, et al. Sex-biased evolutionary forces shape genomic patterns of human diversity. PLoS Genet. 2008;4:e1000202. doi: 10.1371/journal.pgen.1000202. doi:10.1371/journal.pgen.1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gottipati S, Arbiza L, Siepel A, et al. Analyses of X-linked and autosomal genetic variation in population-scale whole genome sequencing. Nat. Genet. 2011;43:741–743. doi: 10.1038/ng.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rice WR. Sex chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 64.Mank JE. Sex chromosomes and the evolution of sexual dimorphism: lessons from the genome. Am. Naturalist. 2009;173:141–150. doi: 10.1086/595754. [DOI] [PubMed] [Google Scholar]
- 65.Cox RM, Calsbeek R. Sexually antagonistic selection, sexual dimorphism, and the resolution of intralocus sexual conflict. Am. Naturalist. 2009;173:176–187. doi: 10.1086/595841. [DOI] [PubMed] [Google Scholar]
- 66.Gibson JR, Chippindale AK, Rice WR. The X chromosome is a hot spot for sexually antagonistic fitness variation. Proc. Roy. Soc. Lond. B. 2002;269:499–505. doi: 10.1098/rspb.2001.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fry JD. The genomic location of sexually antagonistic variation: some cautionary comments. Evolution. 2010;64:1510–1516. doi: 10.1111/j.1558-5646.2009.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Innocenti P, Morrow EH. The sexually antagonistic genes of Drosophila melanogaster. PLoS Biol. 2010;8:e1000335. doi: 10.1371/journal.pbio.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Foerster FK, Coulson T, Sheldon BC, et al. Sexually antagonistic genetic variation for fitness in red deer. Nature. 2007;447:1107–1110. doi: 10.1038/nature05912. [DOI] [PubMed] [Google Scholar]
- 70.Haldane JBS. A mathematical theory of natural and artificial selection. Part 1. Trans. Camb. Philo. Soc. 1924;23:19–41. [Google Scholar]
- 71.Charlesworth B, Coyne JA, Barton N. The relative rates of evolution of sex chromosomes and autosomes. Am. Naturalist. 1987;130:113–146. [Google Scholar]
- 72.Singh ND, Larracuente AM, Clark AG. Contrasting the efficacy of selection on the X and autosomes in Drosophila. Mol. Biol. Evol. 2008;25:454–476. doi: 10.1093/molbev/msm275. [DOI] [PubMed] [Google Scholar]
- 73.Charlesworth B. The effects of deleterious mutations at linked sites. Genetics. 2012;190:5–22. doi: 10.1534/genetics.111.134288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marias G, Charlesworth B. Genome evolution: recombination speeds up adaptive evolution. Curr. Biol. 2003;13:68–70. doi: 10.1016/s0960-9822(02)01432-x. [DOI] [PubMed] [Google Scholar]
- 75.Jaenike J. Sex chromosome meiotic drive. Ann. Rev. Ecol. Syst. 2001;32:25–49. [Google Scholar]
- 76.Frank SA. Divergence of meiotic drive suppression systems as an explanation for sex-biased hybrid sterility and inviability. Evolution. 1991;45:262–267. doi: 10.1111/j.1558-5646.1991.tb04401.x. [DOI] [PubMed] [Google Scholar]
- 77.Hurst LD, Pomiankowski A. Causes of sex-ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane’s rule and related phenomena. Genetics. 1991;128:841–858. doi: 10.1093/genetics/128.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McDermott SR, Noor MAF. The role of meiotic drive in hybrid male sterility. Phil. Trans. Roy. Soc. Lond. B. 2010;365:1265–1272. doi: 10.1098/rstb.2009.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bull JJ. Evolution of Sex Determining Mechanisms. Benjamin Cummings; Menlo Park, CA: 1983. [Google Scholar]
- 80.Charlesworth B. The evolution of sex chromosomes. Science. 1991;251:1030–1033. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- 81.Rice WR. Evolution of the Y sex chromosome in animals. BioScience. 1996;46:331–343. [Google Scholar]
- 82.Wilson MA, Makova KD. Genomic analyses of sex chromosome evolution. Ann. Rev. Genom. Hum. Genet. 2009;10:333–354. doi: 10.1146/annurev-genom-082908-150105. [DOI] [PubMed] [Google Scholar]
- 83.Peck JR. A ruby in the rubbish: beneficial mutations, deleterious mutations, and the evolution of sex. Genetics. 1994;137:594–606. doi: 10.1093/genetics/137.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Otto SP, Pannell JR, Piechel CL, et al. About PAR: the distinct evolutionary dynamics of the pseudoautsomal region. Trends Genet. 2011;27:358–367. doi: 10.1016/j.tig.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 85.Delph LF, Arntz AM, Scotti-Saintagne C, Scotti I. The genomic architecture of sexual dimorphism in thedioecious plant Silene latifolia. Evolution. 64:2873–2886. doi: 10.1111/j.1558-5646.2010.01048.x. [DOI] [PubMed] [Google Scholar]
- 86.Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- 87.Nam K, Ellegren H. The chicken (Gallus gallus) Z chromosome contains at least three nonlinear evolutionary strata. Genetics. 2008;180:1131–1136. doi: 10.1534/genetics.108.090324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bergero R, Forrest A, Kanau E, Charlesworth D. Evolutionary strata of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics. 2007;175:1945–1954. doi: 10.1534/genetics.106.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bachtrog D. A dynamic view of sex chromosome evolution. Curr. Opin. Genet. Devel. 2006;16:578–585. doi: 10.1016/j.gde.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 90.Carvalho AB, Koerich LB, Clark AG. Origin and evolution of Y chromosomes: Drosophila tales. Trends Genet. 2009;25:270–277. doi: 10.1016/j.tig.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Waters PD, Duffy B, Frost CJ, et al. The human Y chromosome derives largely from a single autosomal region added to the sex chromosomes 80–130 million years ago. Cytogenet. Cell Genet. 2001;92:74–79. doi: 10.1159/000056872. [DOI] [PubMed] [Google Scholar]
- 92.Bachtrog D, Hom E, Wong KM, et al. Genomic degradation of a young Y chromosome in Drosophila miranda. Genome Biol. 2008;9:R30. doi: 10.1186/gb-2008-9-2-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Korerich LB, Wang X, Clark AG, Carvalho AB. Low conservation of gene content in the Drosophila Y chromosome. Nature. 2008;456:949–951. doi: 10.1038/nature07463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chippindale AK, Rice WR. Y chromosome polymorphism is a strong determinant of male fitness in Drosophila melanogaster. Proc. Natl. Acad. Sci. 2001;98:5677–5682. doi: 10.1073/pnas.101456898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rohmer C, David JR, Moreteau B, Joly D. Heat-induced male sterility in Drosophila melanogaster. Adaptive genetic variations among geographic populations and role of the Y chromosome. J. Exp. Biol. 2004;207:2735–2742. doi: 10.1242/jeb.01087. [DOI] [PubMed] [Google Scholar]
- 96.Lemos B, Araripe LO, Hartl DL. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science. 2008;319:91–93. doi: 10.1126/science.1148861. [DOI] [PubMed] [Google Scholar]
- 97.Lemos B, Branco A, Hartl DL. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc. Natl. Acad. Sci. 2010;107:15826–15831. doi: 10.1073/pnas.1010383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jiang P-P, Hartl DL, Lemos B. Y Not a dead end: epistatic interactions between Y-linked polymorphisms and genetic background affect global gene expression in Drosophila melanogaster. Genetics. 2010;186:109–118. doi: 10.1534/genetics.110.118109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Paredes S, Branco AT, Hartl DL, et al. Ribosomal DNA deletions modulate genome-wide gene expression: “rDNA-sensitive” genes and natural variation. PLoS Genet. 2011;7:e1001376. doi: 10.1371/journal.pgen.1001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ezaz T, Stiglec R, Veyrunes F, Graves JAM. Relationships between vertebrate ZW and XY sex chromosome systems. Curr. Biol. 2006;16:R736–R743. doi: 10.1016/j.cub.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 101.Bellott DW, Skaletsky H, Pyntikova T, et al. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature. 2010;466:612–617. doi: 10.1038/nature09172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Traut W, Sahara K, Martec F. Sex chromosomes and sex determination in Lepidopetra. Sex. Dev. 2008;1:332–346. doi: 10.1159/000111765. [DOI] [PubMed] [Google Scholar]
- 103.Sturtevant AH. Linkage in the silkworm moth. Am. Naturalist. 1915;48:315–317. [Google Scholar]
- 104.Pease JB, Hahn MW. Sex chromosomes evolved from independent ancestral linkage groups in winged insects. Mol. Biol. Evol. 2012 doi: 10.1093/molbev/mss010. (in press) [DOI] [PubMed] [Google Scholar]
- 105.Toups MA, Pease JB, Hahn MW. No excess gene movement is detected off the avian or Lepidopteran Z chromosome. Genome Biol. Evol. 2011;3:1381–1390. doi: 10.1093/gbe/evr109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pala I, Naurin S, Stervander M, et al. Evidence of a neo-sex chromosome in birds. Heredity. 2012;108:264–272. doi: 10.1038/hdy.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Parisi M, Nuttall R, Naiman D, et al. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ranz JM, Castilllo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- 109.Sturgill D, Zhang Y, Parisi M, Oliver B. Demasculinization of the X chromosome in the Drosophila genus. Nature. 2007;450:238–241. doi: 10.1038/nature06330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vicoso B, Charlesworth B. The deficit of male-biased genes on the D. melanogaster X chromosome is expression-dependent: a consequence of dosage compensation? J. Mol. Evol. 2009;68:576–583. doi: 10.1007/s00239-009-9235-4. [DOI] [PubMed] [Google Scholar]
- 111.Baker DA, Russell S. Role of testes-specific gene expression in sex-chromosome evolution of Anopheles gambiae. Genetics. 2011;189:1117–1120. doi: 10.1534/genetics.111.133157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Prince EG, Kirkland D, Demuth JP. Hyperexpression of the X chromosome in both sexes results in extensive female bias of X-linked genes in the flour beetle. Genome Biol. Evol. 2010;2:336–346. doi: 10.1093/gbe/evq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromsome inactivation. Nat. Genet. 2004;36:642–646. doi: 10.1038/ng1368. [DOI] [PubMed] [Google Scholar]
- 114.Wu C-I, Xu EY. Sexual antagonism and X inactivation: the SAXI hypothesis. Trends Genet. 2003;19:243–247. doi: 10.1016/s0168-9525(03)00058-1. [DOI] [PubMed] [Google Scholar]
- 115.Zhang YE, Vibranovski MD, Landback P, et al. Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biol. 2010;8:e1000494. doi: 10.1371/journal.pbio.1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Arunkumar KP, Mita K, Nagaraju J. The silkworm Z chromosome is enriched in testis-specific genes. Genetics. 2009;182:493–501. doi: 10.1534/genetics.108.099994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bateson W. Heredity and variation in modern lights. In: Seward AC, editor. Darwin and Modern Science. Cambridge University Press; Cambridge: 1909. pp. 85–101. [Google Scholar]
- 118.Dobzhansky Th. Genetics and the Origin of Species. Columbia University Press; New York: 1937. [Google Scholar]
- 119.Muller HJ. Isolating mechanisms, evolution, and temperature. Biol. Symp. 1942;6:71–125. [Google Scholar]
- 120.Masly JP, Jones CD, Noor MAF, et al. Gene transposition as a cause of hybrid sterility in Drosophila. Science. 2006;313:1448–1450. doi: 10.1126/science.1128721. [DOI] [PubMed] [Google Scholar]
- 121.Orr HA. The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics. 1995;139:1805–1813. doi: 10.1093/genetics/139.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Moyle LC, Nakazato T. Hybrid Incompatibility ‘snowballs’ between Solanum species. Science. 2010;329:1521–1523. doi: 10.1126/science.1193063. [DOI] [PubMed] [Google Scholar]
- 123.Matute DR, Butler IA, Turossini DA, Coyne JA. A test of the snowball theory for the rate of evolution of hybrid incompatibilities. Science. 2010;329:1518–152. doi: 10.1126/science.1193440. [DOI] [PubMed] [Google Scholar]
- 124.Lachance JL, Johnson NA, True JR. The population genetics of X-autosome synthetic lethals and steriles. Genetics. 2011;189:1011–1027. doi: 10.1534/genetics.111.131276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kelly JK, Noor MAF. Speciation by reinforcement: a model derived from studies of Drosophila. Genetics. 1996;143:1485–1497. doi: 10.1093/genetics/143.3.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Good JM, Handel MA, Nachman MW. Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evolution. 2008;62:50–65. doi: 10.1111/j.1558-5646.2007.00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tang S, Presgraves DC. Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science. 2009;323:779–782. doi: 10.1126/science.1169123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cattani MV, Presgraves DC. Genetics and lineage-specific evolution of a lethal hybrid incompatibility between Drosophila mauritiana and its sibling species. Genetics. 2009;181:1545–1555. doi: 10.1534/genetics.108.098392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cattani MV, Presgraves DC. Incompatibility between X chromosome factors causes lethality in hybrids between Drosophila melanogaster and its sibling species. Genetics. 2012;191:549–559. doi: 10.1534/genetics.112.139683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lachance J, True JR. X-autosome incompatibilities in Drosophila melanogaster: tests of Haldane’s rule and geographic patterns within species. Evolution. 2010;64:3035–3048. doi: 10.1111/j.1558-5646.2010.01028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Demuth JP, Wade MJ. Population differentiation in the beetle Tribolium castaneum II: Haldane’s rule and incipient speciation. Evolution. 2007;61:694–699. doi: 10.1111/j.1558-5646.2007.00049.x. [DOI] [PubMed] [Google Scholar]
- 132.Orr HA. A mathematical model of Haldane’s rule. Evolution. 1993;47:1606–1611. doi: 10.1111/j.1558-5646.1993.tb02179.x. [DOI] [PubMed] [Google Scholar]
- 133.Turelli M, Orr HA. The dominance theory of Haldane’s rule. Genetics. 1995;140:389–402. doi: 10.1093/genetics/140.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Turelli M, Orr HA. Dominance, epistasis, and the genetics of postzygotic isolation. Genetics. 2000;154:1663–1679. doi: 10.1093/genetics/154.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Turelli M, Begun DJ. Haldane’s rule and X-chromosome size in Drosophila. Genetics. 1997;147:1799–1815. doi: 10.1093/genetics/147.4.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Koevoets T, Beukeboom LW. Genetics of postzygotic isolation and Haldane’s rule in haplodiploids. Heredity. 102:16–23. doi: 10.1038/hdy.2008.44. [DOI] [PubMed] [Google Scholar]
- 137.Sawamura 1996.
- 138.Wu CI. A note on Haldane’s rule: hybrid inviability versus hybrid sterility. Evolution. 1992;46:1584–1587. doi: 10.1111/j.1558-5646.1992.tb01152.x. [DOI] [PubMed] [Google Scholar]
- 139.Coyne JA, Orr HA. Patterns of speciation in Drosophila” revisited. Evolution. 1997;51:295–303. doi: 10.1111/j.1558-5646.1997.tb02412.x. [DOI] [PubMed] [Google Scholar]
- 140.Sasa MM, Chippindale PT, Johnson NA. Patterns of postzygotic reproductive isolation in frogs. Evolution. 1998;52:1811–1820. doi: 10.1111/j.1558-5646.1998.tb02258.x. [DOI] [PubMed] [Google Scholar]
- 141.Wade MJ, Shuster SM. Don’t throw Bateman out with the bathwater! Integ. Comp. Zool. 2005;45:261–268. doi: 10.1093/icb/45.5.945. [DOI] [PubMed] [Google Scholar]
- 142.Torgerson DG, Kulathinal RJ, Singh RS. Mammalian sperm proteins are rapidly evolving: evidence of positive selection in functionally diverse genes. Mol. Biol. Evol. 2002;19:1973–1980. doi: 10.1093/oxfordjournals.molbev.a004021. [DOI] [PubMed] [Google Scholar]
- 143.Haerty W, Jagadeeshan S, Kulathinal RJ, et al. Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics. 2007;177:1321–1335. doi: 10.1534/genetics.107.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Phandis N, Orr HA. A single gene causes both male sterility and segregation distorter in Drosophila hybrids. Science. 2009;323:376–379. doi: 10.1126/science.1163934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Phandis N. Genetic architecture of male sterility and segregation distortion in Drosophila pseudoobscura Bogota-USA hybrids. Genetics. 2011;189:1001–1009. doi: 10.1534/genetics.111.132324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ortiz-Barrientos D, Counterman BA, Noor MAF. Gene expression divergence and the origin of hybrid dysfunctions. Genetica. 2007;129:71–81. doi: 10.1007/s10709-006-0034-1. [DOI] [PubMed] [Google Scholar]
- 147.Good JM, Giger T, Dean MD, Nachman MW. Widespread over-expression of the X chromosome in sterile F1 hybrid mice. PLoS Genet. 2010;6:e1001148. doi: 10.1371/journal.pgen.1001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Haerty W, Singh RS. Gene regulation divergence is a major contributor to the Dobzhansky-Muller incompatibilities seen between species of Drosophila. Mol. Biol. Evol. 2006;23:1707–1714. doi: 10.1093/molbev/msl033. [DOI] [PubMed] [Google Scholar]
- 149.Moehring AJ, Teeter KC, Noor MAF. Genome-wide patterns of expression in Drosophila pure species and hybrid males. II. Examination of multiple-species hybridizations, platforms, and life cycle stages. Mol. Biol. Evol. 2006;24:137–147. doi: 10.1093/molbev/msl142. [DOI] [PubMed] [Google Scholar]
- 150.Ting C-T, Tsaur S-C, Wu M-L, Wu C-I. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science. 1998;282:1501–1504. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- 151.Lu X, Shapiro JA, Ting C-T, et al. Genome-wide misexpression of X-linked versus autosomal genes associated with hybrid male sterility. Genome Res. 2010;20:1097–1102. doi: 10.1101/gr.076620.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bayes JJ, Malik HS. Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science. 2009;326:1538–1541. doi: 10.1126/science.1181756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Johnson NA, Hollocher H, Noonberg E, Wu C-I. The effects of interspecific Y chromosome replacements on hybrid sterility within the Drosophila simulans clade. Genetics. 1993;134:251–260. doi: 10.1093/genetics/135.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sackton TB, Montenegro H, Hartl DL, Lemos B. Disruption of testis-specific gene expression and male reproductive phenotypes in heterospecific Y chromosome introgressions in Drosophila. Proc. Natl. Acad. Sci. 2011;108:17046–17051. doi: 10.1073/pnas.1114690108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Eo SH, DeWoody JA. Evolutionary rates of mitochondrial genomes correspond to diversification rates and to contemporary species richness in birds and reptiles. Proc. Roy. Soc. Lond. B. 2010;277:3587–3592. doi: 10.1098/rspb.2010.0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Phillips BC, Edmands S. Does the speciation clock tick more slowly in the absence of sex chromosomes? Bioessays. 2012;34:166–169. doi: 10.1002/bies.201100164. [DOI] [PubMed] [Google Scholar]
- 157.Waddington CH. The Strategy of the Genes. George Allen & Unwin; London: 1957. [Google Scholar]
- 158.Jamniczky HA, Boughner JC, Rolian C, et al. Re-discovering Waddington in the post-genomic age. Bioessays. 2010;32:553–558. doi: 10.1002/bies.200900189. [DOI] [PubMed] [Google Scholar]
- 159.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 160.Park Y, Kuroda M. Epigenetic aspects of X chromosome dosage compensation. Science. 2001;293:1083–1085. doi: 10.1126/science.1063073. [DOI] [PubMed] [Google Scholar]
- 161.Jablonka E, Lamb MJ. Epigenetic inheritance in evolution. J. Evol. Biol. 1998;11:159–183. [Google Scholar]
- 162.Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 163.Bachtrog D. Sex chromosome evolution: molecular aspects of Y-chromosome degeneration in Drosophila. Genome Res. 2005;15:1393–1401. doi: 10.1101/gr.3543605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pritham EJ, Zhang YH, Feschotte C, Kesseli RV. Transposable element family with transcriptionally active Y-linked copies in the white campion, Silene latifolia. Genetics. 2003;165:799–807. doi: 10.1093/genetics/165.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Skaletsky H, Kuroda-Kawaguda T, Minx PJ, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–827. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 166.Hughes JF, Skaletsky H, Brown LG, et al. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature. 2012;483:82–86. doi: 10.1038/nature10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rozen S, Marszalek JD, Alagappan RK, et al. Remarkably little variation in proteins encoded by the Y chromosome’s single-copy genes, implying effective purifying selection. Am. J. Hum. Genet. 2009;85:923–928. doi: 10.1016/j.ajhg.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hughes JF, Skaletsky H, Pyntikova T, et al. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature. 2010;463:536–539. doi: 10.1038/nature08700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Shetty S, Kirby P, Zarkower D, Graves JAM. DMRT1 in a ratite bird: evidence for a role in sex determination and discovery of a putative regulatory element. Cyto. Genome Res. 2002;99:245–251. doi: 10.1159/000071600. [DOI] [PubMed] [Google Scholar]
- 170.Ellegren H. Evolution of the avian sex chromosomes and their role in sex determination. Trends Ecol. Evol. 2000;15:188–192. doi: 10.1016/s0169-5347(00)01821-8. [DOI] [PubMed] [Google Scholar]
- 171.Smith CA, Rosezler KN, Ohnesong T, et al. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461:267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]