Abstract
Molybdenum cofactor deficiency (MoCD) is a rare inherited metabolic disorder characterized by severe and progressive neurological damage mainly caused by the loss of sulfite oxidase activity. Elevated urinary levels of sulfite, thiosulfate, and S-sulfocysteine (SSC) are hallmarks in the diagnosis of MoCD and sulfite oxidase deficiency (SOD). Recently, a first successful treatment of a human MoCD type A patient based on a substitution therapy with the molybdenum cofactor precursor cPMP has been reported, resulting in nearly complete normalization of MoCD biomarkers. Knowing the rapid progression of the disease symptoms in nontreated patients, an early diagnosis of MoCD as well as a sensitive method to monitor daily changes in SSC levels, a key marker of sulfite toxicity, is crucial for treatment outcome. Here, we describe a fast and sensitive method for the analysis of SSC in human urine samples using high performance liquid chromatography (HPLC). The analysis is based on precolumn derivatization with O-phthaldialdehyde (OPA) and separation on a C18 reverse phase column coupled to UV detection. The method was extended to human serum analysis and no interference with endogenous amino acids was found. Finally, SSC values from 45 pediatric urine, 75 adult urine, and 24 serum samples from control individuals as well as MoCD patients are reported. Our method represents a cost-effective technique for routine diagnosis of MoCD and SOD, and can be used also to monitor treatment efficiency in those sulfite toxicity disorders on a daily basis.
Introduction
MoCD is a rare inherited metabolic disorder (Johnson et al. 1980; Johnson and Duran 2001) caused by defects in the biosynthesis of the molybdenum cofactor (Moco) leading to the simultaneous loss of activities of all molybdenum-dependent enzymes: sulfite oxidase, xanthine dehydrogenase, aldehyde oxidase and mitochondrial amidoxime-reducing component (mARC) (Schwarz et al. 2009). Affected patients exhibit severe neurological abnormalities, such as microcephaly, seizures, and usually die in early childhood (Johnson and Duran 2001). Sulfite oxidase deficiency (SOD) is less frequent but clinically similar to MoCD, which renders sulfite oxidase as the most important Moco enzyme in humans (Tan et al. 2005). Sulfite oxidase catalyzes the oxidation of sulfite, which is generated throughout the catabolism of sulfur-containing amino acids, to sulfate (Griffith 1987; Johnson and Duran 2001). Xanthine dehydrogenase, another important Moco enzyme, catalyzes the conversion of hypoxanthine to xanthine and further to uric acid (Hille 2005). Aldehyde oxidase catalyzes the hydroxylation of various compounds (Garattini et al. 2009), while mARC was identified as a general detoxifying and pro-drug metabolizing enzyme with yet unknown physiological substrates (Wahl et al. 2010).
Deficiencies of Moco and sulfite oxidase result in the accumulation of sulfite, a highly toxic molecule that breaks disulfide bridges in proteins and cystine, thereby affecting many protein and cellular functions (Zhang et al. 2004). Sulfite first accumulates in liver, where most of the catabolism of sulfur-containing amino acids takes place. Subsequently, accumulation of sulfite in plasma is detectable and finally sulfite crosses the blood–brain-barrier triggering a devastating and progressive neuronal death (Schwarz et al. 2009). Sulfite accumulation is accompanied by the formation of secondary metabolites such as thiosulfate and S-sulfocysteine (SSC) which are common biochemical indicators for MoCD and SOD together with reduced homocysteine levels (Johnson and Duran 2001; Sass et al. 2004). However, isolated SOD can be excluded if the products of other Moco enzymes such as uric acid are reduced in urine and plasma and urinary xanthine and hypoxanthine are elevated. In addition, the absence of urinary urothione as a specific metabolic degradation product of Moco, is a direct indicator for MoCD (Bamforth et al. 1990).
MoCD can be grouped into three types according to the underlying genetic defect. Type A deficiency affects two-thirds of all patients and is caused by a mutation in the MOCS1 gene (Reiss and Johnson 2003). While type A patients lack the first precursor in the biosynthetic pathway of Moco, cyclic pyranopterin monophosphate (cPMP) (Santamaria-Araujo et al. 2004), type B patients accumulate cPMP due to mutations in the MOCS2 gene, which encodes the molybdopterin synthase (Reiss et al. 1999). Up to now only one type C patient with a mutation in the GEPHYRIN gene has been found (Reiss et al. 2001). Using an animal model for human MoCD (Lee et al. 2002), a treatment approach based on a substitution therapy with cPMP has been established for MoCD type A (Schwarz et al. 2004). Recently, a first human exposure to cPMP has been reported in a type A patient with a remarkable normalization of MoCD biomarkers such as sulfite, SSC, xanthine, uric acid, and urothione leading to a significant clinical improvement of the patient (Veldman et al. 2010). This study revealed also the devastating character of the disease, which was manifested by a rapid increase of urinary sulfite, thiosulfate, and SSC values during the first 36 days of life before treatment started. A recent study supported previous data from animal studies and demonstrated that maternal sulfite clearance is able to suppress prenatal brain damage; however, within days after birth a rapid and progressive brain damage with repetitive seizures has occurred (Sie et al. 2010). Therefore, treatment should be initiated as early as possible. Consequently, the need for a fast and sensitive method for an easy diagnosis of MoCD and a daily monitoring of treatment efficiency becomes important.
In the past, reports showed that diagnosis of MoCD is often difficult, as sulfite is very unstable and false-negative results can occur already within four hours of storage of urine (Kutter and Humbel 1969). False-positive results may also occur in the presence of drugs containing reactive sulfhydryl groups such as N-acetylcysteine, mercaptamine, and dimercaprol (Wadman et al. 1983). In contrast, SSC is more stable and suitable for automated analysis and screening. Various methods for determination of SSC were reported, including HPLC-based techniques, electrospray tandem mass spectrometry, and anion exchange chromatography (Demarco et al. 1965; Johnson and Rajagopalan 1995; Pitt et al. 2002). However, only some of these methods provide sufficient sensitivity for the determination of SSC in urine samples, while extended analysis time, complex sample clean-up procedures, or instability of certain SSC derivatives limited their use in an automated, fast, and reproducible setting. A general method currently used for the diagnosis of MoCD is based on a sulfite dipstick for the rapid detection of urinary sulfite. Furthermore, most SSC analyses are performed with the classical amino acid analyzer, but quantification is limited due to the early elution of SSC. In addition, poor specificity and possible interference from other metabolites were also reported by using the amino acid analyzer (Rashed et al. 2005).
Here, we describe a simple, fast, and sensitive HPLC-based method using automated precolumn derivatization with OPA and UV detection at 338 nm for the determination of urinary SSC levels. The method was extended to serum analysis and SSC separation from endogenous amino acids was demonstrated. We reported SSC control levels derived from 45 pediatric urine, 75 adult urine, and 24 adult serum samples. Furthermore, SSC values from MoCD patients are reported.
Materials and Methods
Reagents
HPLC-grade methanol, acetonitrile, and disodium hydrogen phosphate were obtained from BDH Prolabo (VWR International GmbH, Darmstadt, Germany). Sodium tetraborate, boric acid, 5-Sulfosalicylic acid, and S-sulfocysteine were from Sigma–Aldrich (St. Louis, USA). O-Phthaldialdehyde (OPA) was purchased from Alfa Aesar GmbH & Co KG (Karlsruhe, Germany). Preparation of the derivatization reagent was based on the protocol of Jakoby (Jakoby and Griffith 1987) and achieved by dissolving 1 g of OPA in 10 ml methanol, mixed with 90 ml 0.4 M borate buffer pH 10.2 and 400 μl 2-mercaptoethanol. The prepared derivatization reagent was stored in 1 ml aliquots at −20°C.
Chromatographic Conditions
HPLC analyses were carried out on an Agilent 1200 SL system (Agilent Technologies GmbH, Boeblingen, Germany) consisting of a binary pump (SL series), vacuum degasser, autosampler, thermostated column compartment (SL series), diode array detector (SL series), fluorescence detector, all controlled by Agilent ChemStation software. The mobile phase consisted of buffer A (10 mM Na2HPO4; 10 mM Na2B4O7) and eluent B (45% acetonitrile 45% methanol and 10% water). Different reversed-phase BEH (ethylene bridged hybrid)-C18 columns were used: XBridge (50 × 4.6 mm, 2.5 μm, Waters GmbH, Eschborn, Germany) and ACQUITY UPLC (50 × 2.1 mm, 1.7 μm, Waters GmbH, Eschborn, Germany) and separation was carried out isocratically with 10% eluent B. For analysis of all amino acids, an additional gradient from 10 to 60% eluent B was applied to the column after elution of SSC. Precolumn derivatization was carried out using an automated autosampler, which was programmed to mix 9 μl of the sample with 1 μl of derivatization reagent, after an incubation time of 0.2 min, the mixture was injected and the injection valve was bypassed to achieve the derivatization of the next sample, thus eliminating the time needed for the derivatization procedure. Detection was carried out by UV absorbance at 338 nm and compound identification was achieved by comparing the retention time with that obtained for SSC standard. Peak area was used for calibration. SSC amount was determined by standard addition and normalized to creatinine in urine samples.
Samples and Standard Preparation
Urine samples were centrifuged for 15 min at 13,000 × g, the supernatant was filtered through a 0.2-μm filter and no additional solid phase extraction was performed. Urine samples were diluted in HPLC-grade water and directly analyzed. For the analysis of serum samples, fresh blood samples were centrifuged for 10 min at 1,300 × g and serum was collected and stored at −20°C until further analysis. For serum protein precipitation, 100 μl 5% 5-sulfosalicylic acid were added to 200 μl serum and 200 μl HPLC-grade water. Precipitated proteins were removed by centrifugation for 15 min at 13,000 × g. The supernatant fraction was filtered through a 0.2-μm filter and directly analyzed.
Creatinine Analysis
Creatinine determination was based on the Jaffe method (Vasiliades 1976; Kroll et al. 1984). Briefly, 50 μl of diluted urine samples were mixed with 150 μl alkaline picrate solution (1.2% picric acid in 0.75 M sodium hydroxide). After an incubation time of 15 min, the formation of an orange-red complex between creatinine and alkaline picrate was quantified by measuring the absorbance at 490 nm using a microplate reader (BioTek, Friedrichshall, Germany). Method calibration was achieved by measuring the absorbance of different creatinine standard solutions.
Validation Procedure: Linearity and Calibration
Calibration of the method was carried out using three different conditions. SSC standard solutions were added to water, SSC-free urine, and serum samples. Peak area was used for quantification and linearity was determined by using a wide SSC concentration range from 2.5 to 200 μM.
Validation Procedure: Specificity
SSC determination was carried out using isocratic elution from the HPLC column. Under those conditions, the majority of the amino acids were eluted after the SSC peak. To demonstrate this, a standard solution containing all primary amino acids including SSC and taurine was separated. The specificity of the method was also determined by analyzing urine and serum samples under the same chromatographic conditions to show separation of SSC from endogenous amino acids.
Validation Procedure: Limit of Detection and Limit of Quantification
Limit of detection (LOD) and limit of quantification (LOQ) were determined by measuring the standard deviation of peak area and slope of the calibration curve. LOD and LOQ were determined by adding increasing amounts of SSC standard in the range of 2.5–200 μM to water, SSC-free urine, and serum samples.
Validation Procedure: Precision and Reproducibility
Intraday precision was determined by analyzing samples three times a day randomly and interday precision was determined by analyzing samples at ten different days. Relative standard deviation (RSD) of peak area was used and analysis was carried out at three different concentrations. Low quantity control (LQC), median quantity control (MQC), and high quantity control (HQC) samples were prepared by spiking, water, SSC-free urine, and serum samples to a final concentration of 10, 60, and 200 μM, respectively. Reproducibility of the method was assayed by using different matrix batches, different column diameters, and lengths. Different separation methods were tested including isocratic or gradient elution at different flow rates.
Validation Procedure: Recovery
The percentage of recovery was determined at three concentration levels LQC, MQC, and HQC. SSC-free urine and serum samples were spiked with SSC standard. Peak area of the response was compared to that of standard solutions prepared in water.
Results and Discussion
Derivatization
HPLC analysis of amino acids with OPA derivatization is one of the most sensitive methods for amino acids quantification with detection limits in the femtomole range. OPA reacts with all primary amino acids under alkaline conditions and in the presence of thiols OPA forms iso-indoles that can be detected either by fluorescence (excitation 230 nm, emission 450 nm) or absorbance at 340 nm (Hill et al. 1979). Amino acids carrying a free thiol group such as cysteine or homocysteine cannot be directly detected with OPA derivatization and a modification prior to the derivatization reaction is needed. Carboxymethylation is usually used to convert cysteine to carboxymethylcysteine prior to OPA derivatization, thus leading to a fluorescence intensity similar to other amino acids (Jarrett et al. 1986). Based on the chemical structure of SSC lacking a free thiol group, we have chosen to use a direct derivatization of SSC with OPA without prior modification.
Chromatography
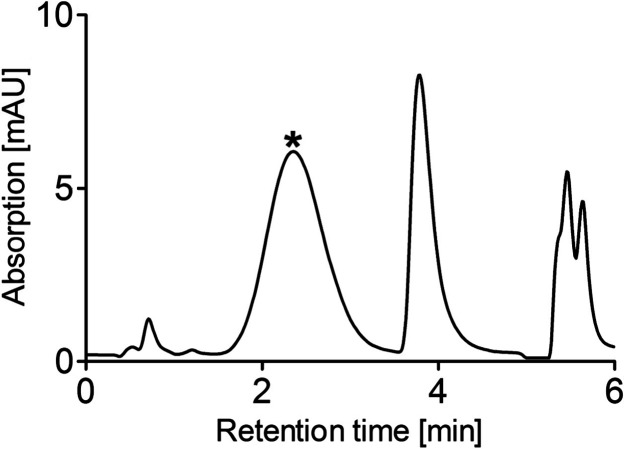
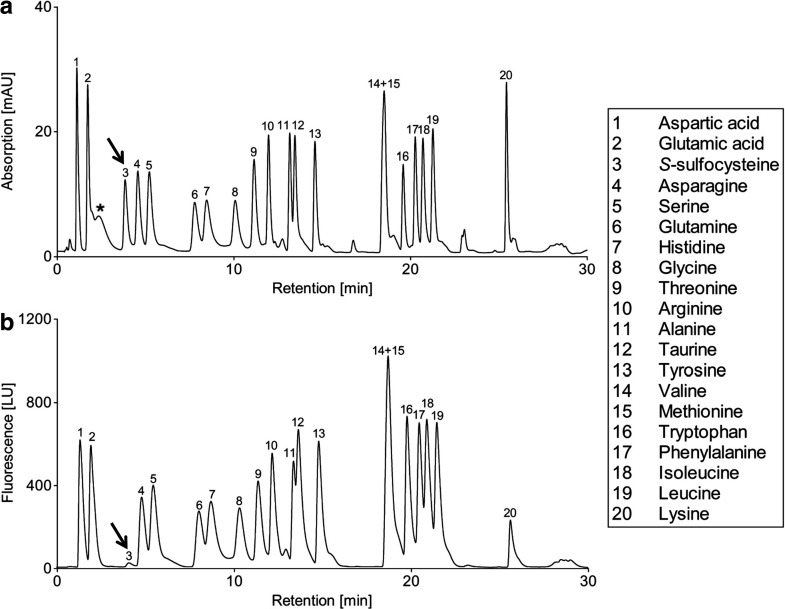
One of the critical steps in the OPA derivatization procedure is the instability of the amino acid derivatives, which is manifested by the decay of fluorescence over time (Hogan et al. 1982; Fleury and Ashley 1983; Cooper et al. 1984). To overcome this problem, we aimed to develop fast chromatographic methods using short columns (2.1 mm × 50 mm) and UPLC particles (reversed phase C18 1.7 μm) to achieve high resolution at short analysis time. As a result, separation was completed within 6 min using isocratic elution allowing rapid analysis. Under the chromatographic conditions described, SSC yielded a sharp peak eluting at 3.8 min (Fig. 1). Specificity of the method was determined by analyzing a standard solution containing all primary amino acids including SSC and taurine and a gradient elution was applied after elution of SSC. The results showed that most of the amino acids are eluted after the SSC peak, and thus the chromatographic conditions optimized for SSC allowed a clear separation from all other amino acids (Fig. 2a).
Fig. 1.
HPLC analysis of standard SSC solution. Twenty micromolar SSC standard was prepared in H2O and analyzed under chromatographic conditions described in Materials and Methods. Retention time of SSC peak was 3.8 min. The large peak eluting before SSC (depicted by an asterisk) represents a degradation product of OPA. The OPA excess is used to ensure quantitative derivatization of all primary amino acids in urine and serum
Fig. 2.
HPLC analysis of amino acids. A standard solution of 20 μM containing all primary amino acids and additionally SSC and taurine were prepared in H2O and separated under chromatographic conditions described in Materials and Methods. Detection was carried out by UV absorbance at 338 nm (a) and fluorescence (b). All amino acids are numbered and shown in the legend. The SSC peak is depicted by an arrow in both panels. The differences in peak shape resulted from the different elution methods used: For the first eight amino acids isocratic elution was used while the remaining 12 amino acids were eluted with a gradient from 10 to 60% of eluent B
Detection of OPA derivatives is carried out by using either fluorescence or UV absorbance. However, SSC showed a very low specific fluorescence when compared to other primary amino acids (Fig. 2b). Furthermore, all primary amino acids and SSC OPA-derivatives showed a similar absorbance ratio at 338 nm; thus, using the UV approach a direct identification of accumulated OPA derivatives can be easily achieved.
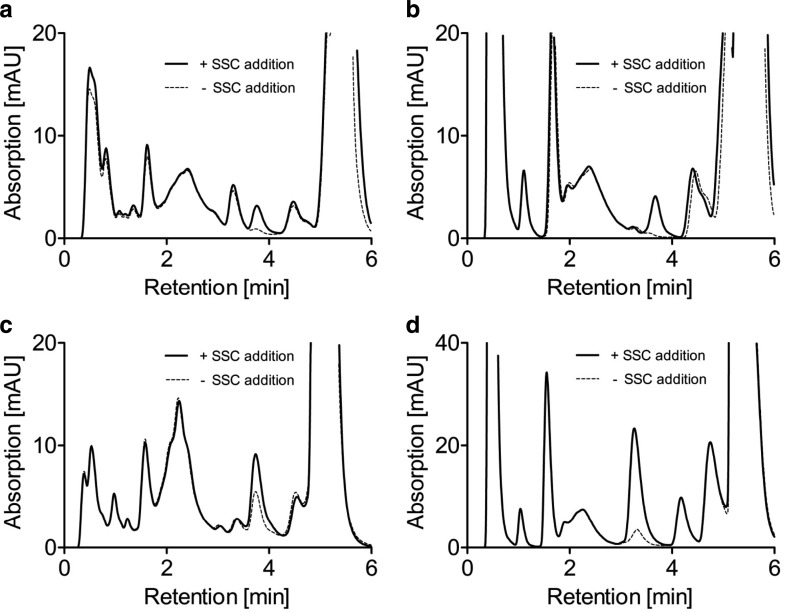
We analyzed urine and serum samples derived from control individuals for SSC content and the SSC peak was separated (Fig. 3a, b). The SSC peak was identified by comparing its retention time to that of a separately injected standard peak and peak purity was proven by spectral homogeneity (data not shown). SSC concentration was determined by standard addition (Fig. 3a, b). Analysis of samples derived from healthy individuals revealed the presence of SSC in urine, while in serum SSC was below detection limit. In contrast, analysis of urine and serum samples derived from MoCD patients showed an accumulated SSC peak, which was quantified in both urine and serum samples (Fig. 3c, d).
Fig. 3.
Standard addition method for quantification of SSC in urine and serum. Control urine (a) and control serum (b) was analyzed and the standard addition method was used to quantify SSC. Control urine sample shows an SSC peak close to the detection limit whereas in control serum SSC was not detectable. Analysis of urine (c) and serum (d) samples derived from non-treated MoCD patient show a clear accumulation of SSC. All chromatograms show an overlay of two analyses. The first analysis was carried out by diluting samples with H2O (−SSC addition) while prior to the second analysis the samples were diluted with SSC standard (+SSC addition). SSC amounts were quantified by comparing peak area in both analyses and SSC amounts were normalized to creatinine for urine samples and expressed in mmol/mol creatinine
HPLC analysis of OPA-derivatized amino acids is usually carried out using alkaline buffers, which provide a greater stability of the amino acid derivatives (Cooper et al. 1984). However, a major problem of alkaline running buffers is the short lifetime of silica-based columns. The pH of the mobile phase used under our chromatographic conditions was not adjusted and corresponded to pH 9.2. Using a conventional silica-based C18 reversed phase column, we observed a continuous decrease in the retention time of the SSC derivative with increasing number of injections (data not shown). After 500 injections, the column completely lost its separation performance. The sensitivity against alkaline buffers was resolved by using Bridged Ethyl Hybrid (BEH) particles (Waters, Germany). The BEH particles consist of modified silica particles that provide increased pH stability in the range of pH 1–12 compared to the conventional silica-based C18 resins with a maximal suitable pH of 8.0. Using the BEH C18 silica-based columns, we did not observe any change in the retention time of any analyzed derivatives for more than 5,000 injections.
Calibration, Precision, and Recovery
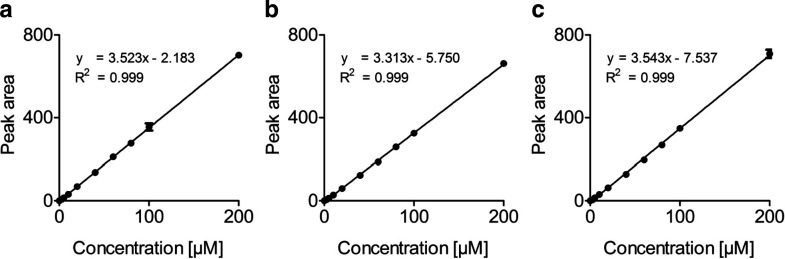
Method calibration was performed using ten different concentrations of SSC standard ranging from 0 to 200 μM. All calibration standards were run in triplicate and peak area was used for quantification. Linearity of the method was determined in three different experimental set-ups. Different concentrations of standard solutions were prepared in (1) water, (2) urine, or (3) serum and subsequently analyzed. The calibration curves displayed high linearity over the concentration range investigated (Fig. 4). LOD ranged from 1.08 to 1.44 μM and LOQ ranged from 3.27 to 4.38 for serum and urine analysis, respectively (Table 1).
Fig. 4.
Liniarity of SSC quantification. Ten SSC concentrations ranging from 0 to 200 μM were spiked in H2O (a), SSC-free urine (b) and SSC-free serum (c) and subsequently used for method calibration. All samples were run in triplicates and points represent means ± relative standard deviations (RSD) of peak area. Equation and correlation coefficient (R2) are given for the linear regression curves of the three different conditions
Table 1.
Method sensitivity, precision and recovery of SSC
| Specimen | LOD (μM) | LOQ (μM) | Intraday stability RSD (%) | Interday stability RSD (%) | Recovery (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LQC | MQC | HQC | LQC | MQC | HQC | LQC | MQC | HQC | |||
| H2O | 1.18 | 3.58 | 1.12 | 3.86 | 2.19 | 3.34 | 4.15 | 0.57 | – | – | – |
| Urine | 1.44 | 4.38 | 0.76 | 1.45 | 0.81 | 8.22 | 1.97 | 6.65 | 96.10–97.73 | 99.60–100.75 | 96.09–97.48 |
| Serum | 1.08 | 3.27 | 0.70 | 1.04 | 2.77 | 0.56 | 3.87 | 3.40 | 95.77–96.72 | 92.16–93.53 | 98.89–102.84 |
Derivatization methods add extra steps in the sample preparation procedure resulting often in variations between repeated injections. By using automated precolumn derivatization these variations can be reduced to a minimum. Intraday and interday precision of the method were determined and confirmed the low variation in repeated sample injections (Table 1). The intraday and interday precision of the method ranged from 0.70 to 3.86% and 0.56 to 8.22%, respectively. Recovery was determined for urine and serum at three concentration levels of SSC and ranged from 87.34 to 94.76% for urine and 92.16 to 102.84% for serum (Table 1).
Analysis of Human Samples
One of the applications of the described method is the fast diagnostic analysis of urine samples derived from individuals with suspected MoCD or SOD. A positive sulfite dipstick is the starting point of diagnostic tests commonly used in clinics and most laboratories. A typical diagnostic procedure usually includes two steps: (1) fresh urine sample is analyzed and SSC peak is identified by comparing the retention time with that of standard and (2) standard addition verifies the identity of the peak and quantifies SSC. For this, a defined amount of SSC was added to urine or serum sample, and the resulting chromatogram was compared to the chromatogram obtained without standard addition. A positive control displayed a symmetric SSC Peak with an increased area (Fig. 3c, d).
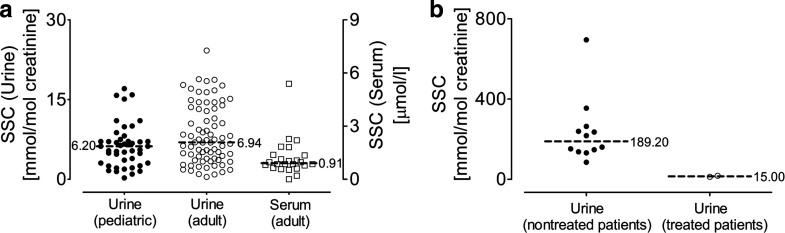
Here, the method was applied to the analysis of 45 pediatric urine, 75 adult urine, and 24 serum samples all derived from control individuals. SSC was detected in all analyzed urine samples demonstrating the low detection limit of the method (Fig. 5a). The values in pediatric urine samples ranged from 0.26 to 18.83 mmol/mol creatinine (median: 6.20 mmol/mol creatinine), and SSC levels of adult urine samples were comparable with a median of 6.94 mmol/mol creatinine. This finding demonstrates a similar SSC excretion level in both age groups. Serum analysis revealed in general low SSC levels and the values ranged from 0 to 5.39 μmol/l with a median of 0.91 (Fig. 5a). This finding suggests a continuous excretion of SSC in urine without any threshold barrier.
Fig. 5.
SSC levels in control individuals and MoCD patients. SSC was quantified in urine and serum samples derived from control individuals (a) and urine samples from MoCD patients (b). In total, 45 pediatric urine samples (filled circles), 75 adult urine (empty circles) and 24 adult serum samples (empty squares) were analyzed to determine the SSC threshold in control individuals. SSC values of urine samples derived from 12 nontreated MoCD patients are shown in panel b (filled circles) and from two cPMP-treated MoCD patients (empty circles). SSC amount is given in mmol/mol creatinine and μmol/l for urine and serum samples, respectively. Median values are shown by horizontal bars
Different urine samples from nontreated MoCD patients were analyzed and SSC was detectable from the first days of life. SSC analysis of urine samples derived from 12 genetically confirmed MoCD type A or type B individuals showed SSC levels ranging from 85 to 695 mmol/mol creatinine (Fig. 5b). Except the index case (Veldman et al. 2010), all currently cPMP-treated MoCD patients (Schwarz et al., unpublished results) are monitored for their SSC levels using the here described method. Here, we show average data over a six months period for two patients treated with cPMP (Schwahn et al. 2010) (Fig. 5b).
Pitt and colleagues reported SSC urinary levels of 0–9 mmol/mol creatinine for control individuals (Pitt et al. 2002) with a median of approximately 2 mmol SSC/mol creatinine, which are lower than the SSC urinary levels measured in this study with a median of 6.94 mmol/mol creatinine. However, the authors reported a poor recovery of SSC in low controls by using MS in negative-ion mode, which they attribute to high salt content of urine samples and the fact that a single internal standard was used. We did not observe any vulnerability of the method used in this study to the ionic content of the sample. Furthermore, we quantified SSC by the use of an identical internal standard. In addition, the reported SSC urinary values of a cPMP-treated MoCD type A patient (Veldman et al. 2010) as well as the reported values of two cPMP-treated MoCD patients in this study are in the same range as the values determined for healthy individuals. Taking into account that urine analysis was carried out without precleaning procedures, thus maximizing recovery, we conclude that the urinary SSC values presented in this study can serve as reference values, which should help to define a threshold limit for future routine diagnostics.
Conclusion
We present a simple, fast, and robust method for the determination of SSC in human urine. No sample clean-up was required, thus reducing analysis time and the risk of contamination. We have shown that our method can be extended to the determination of serum SSC levels with the inclusion of an additional precipitation step. High sensitivity of the method enabled determination of urinary SSC levels in control pediatric and adult individuals revealing a similar excretion range with a median of 6.20 and 6.94 mmol/mol creatinine, respectively. The method has been successfully used for the diagnosis of MoCD/SOD and routine monitoring of cPMP-treated MoCD patients (Schwarz et al., unpublished results). The sensitivity, linearity, and accuracy of the HPLC derivatization method demonstrated in this study, together with the short analysis time, makes this method suitable for automated diagnostic analysis which can be used in a multi-well plate-based autosampler. Given the fact that MoCD is still considered an underdiagnosed inborn error in metabolism (Johnson and Duran 2001), the described method should provide diagnostic laboratories with an accurate, fast, and cost-effective method to quantify SSC levels in urine and serum.
Acknowledgments
We thank Parmanand Arjune, MD, PhD (Cologne, Germany) for providing a large number of urine and serum samples. We are grateful to James Pitt, PhD (Melbourne, Australia) for the comparative analysis of selected urine samples. We gratefully acknowledge Bernd Schwahn, MD, Steve Bowhay, PhD, Peter G. Galloway, MD (Royal Hospital for Sick Children, Glasgow, Scotland), Julia Hennemann, MD (Pediatric Unit, Charité Berlin, Germany), Francjan J. van Spronsen, MD, PhD (University Medical Center, Groningen, Netherlands), and Illona Weis, MD (Pediatric Unit, Gemeinschaftsklinikum Koblenz-Mayen, Germany) and other colleagues for providing urine samples of patients.
Abbreviations
- HPLC
High performance liquid chromatography
- MoCD
Molybdenum cofactor deficiency
- Moco
Molybdenum cofactor
- OPA
O-phthaldialdehyde
- SOD
Sulfite oxidase deficiency
- SSC
S-sulfocysteine
Footnotes
Competing interests: None declared.
References
- Bamforth FJ, Johnson JL, Davidson AG, Wong LT, Lockitch G, Applegarth DA. Biochemical investigation of a child with molybdenum cofactor deficiency. Clin Biochem. 1990;23:537–542. doi: 10.1016/0009-9120(90)80046-L. [DOI] [PubMed] [Google Scholar]
- Cooper JD, Ogden G, McIntosh J, Turnell DC. The stability of the o-phthalaldehyde/2-mercaptoethanol derivatives of amino acids: an investigation using high-pressure liquid chromatography with a precolumn derivatization technique. Anal Biochem. 1984;142:98–102. doi: 10.1016/0003-2697(84)90522-0. [DOI] [PubMed] [Google Scholar]
- Demarco C, Mosti R, Cavallin D. Column chromatography of some sulfur-containing amino acids. J Chromatogr. 1965;18:492. doi: 10.1016/S0021-9673(01)80406-4. [DOI] [PubMed] [Google Scholar]
- Fleury MO, Ashley DV. High-performance liquid chromatographic analysis of amino acids in physiological fluids: on-line precolumn derivatization with o-phthaldialdehyde. Anal Biochem. 1983;133:330–335. doi: 10.1016/0003-2697(83)90092-1. [DOI] [PubMed] [Google Scholar]
- Garattini E, Fratelli M, Terao M. The mammalian aldehyde oxidase gene family. Hum Genomics. 2009;4:119–130. doi: 10.1186/1479-7364-4-2-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith OW. Mammalian sulfur amino acid metabolism: an overview. Methods Enzymol. 1987;143:366–376. doi: 10.1016/0076-6879(87)43065-6. [DOI] [PubMed] [Google Scholar]
- Hill DW, Walters FH, Wilson TD, Stuart JD. High performance liquid chromatographic determination of amino acids in the picomole range. Anal Chem. 1979;51:1338–1341. doi: 10.1021/ac50044a055. [DOI] [PubMed] [Google Scholar]
- Hille R. Molybdenum-containing hydroxylases. Arch Biochem Biophys. 2005;433:107–116. doi: 10.1016/j.abb.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Hogan DL, Kraemer KL, Isenberg JI. The use of high-performance liquid chromatography for quantitation of plasma amino acids in man. Anal Biochem. 1982;127:17–24. doi: 10.1016/0003-2697(82)90138-5. [DOI] [PubMed] [Google Scholar]
- Jakoby WBE, Griffith OWE. Sulfur and sulfur amino acids. London: Academic; 1987. [Google Scholar]
- Jarrett HW, Cooksy KD, Ellis B, Anderson JM. The separation of o-phthalaldehyde derivatives of amino acids by reversed-phase chromatography on octylsilica columns. Anal Biochem. 1986;153:189–198. doi: 10.1016/0003-2697(86)90079-5. [DOI] [PubMed] [Google Scholar]
- Johnson JL, Duran M, et al. Molybdenum cofactor deficiency and isolated sulfite oxidase deficiency. In: Scriver C, et al., editors. The metabolic and molecular bases of inherited disease. New York: McGraw-Hill; 2001. pp. 3163–3177. [Google Scholar]
- Johnson JL, Rajagopalan KV. An HPLC assay for detection of elevated urinary S-sulphocysteine, a metabolic marker of sulfite oxidase deficiency. J Inherit Metab Dis. 1995;18:40–47. doi: 10.1007/BF00711371. [DOI] [PubMed] [Google Scholar]
- Johnson JL, Waud WR, Rajagopalan KV, Duran M, Beemer FA, Wadman SK. Inborn errors of molybdenum metabolism: combined deficiencies of sulfite oxidase and xanthine dehydrogenase in a patient lacking the molybdenum cofactor. Proc Natl Acad Sci U S A. 1980;77:3715–3719. doi: 10.1073/pnas.77.6.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll MH, Hagengruber C, Elin RJ. Reaction of picrate with creatinine and cepha antibiotics. Clin Chem. 1984;30:1664–1666. [PubMed] [Google Scholar]
- Kutter D, Humbel R. Screening for sulfite oxidase deficiency. Clin Chim Acta. 1969;24:211–214. doi: 10.1016/0009-8981(69)90314-3. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Adham IM, Schwarz G, Kneussel M, Sass JO, Engel W, Reiss J. Molybdenum cofactor-deficient mice resemble the phenotype of human patients. Hum Mol Genet. 2002;11:3309–3317. doi: 10.1093/hmg/11.26.3309. [DOI] [PubMed] [Google Scholar]
- Pitt JJ, Eggington M, Kahler SG. Comprehensive screening of urine samples for inborn errors of metabolism by electrospray tandem mass spectrometry. Clin Chem. 2002;48:1970–1980. [PubMed] [Google Scholar]
- Rashed MS, Saadallah AA, Rahbeeni Z, Eyaid W, Seidahmed MZ, Al-Shahwan S, Salih MA, Osman ME, Al-Amoudi M, Al-Ahaidib L, Jacob M. Determination of urinary S-sulphocysteine, xanthine and hypoxanthine by liquid chromatography-electrospray tandem mass spectrometry. Biomed Chromatogr. 2005;19:223–230. doi: 10.1002/bmc.439. [DOI] [PubMed] [Google Scholar]
- Reiss J, Johnson JL. Mutations in the molybdenum cofactor biosynthetic genes MOCS1, MOCS2, and GEPH. Hum Mutat. 2003;21:569–576. doi: 10.1002/humu.10223. [DOI] [PubMed] [Google Scholar]
- Reiss J, Dorche C, Stallmeyer B, Mendel RR, Cohen N, Zabot MT. Human molybdopterin synthase gene: genomic structure and mutations in molybdenum cofactor deficiency type B. Am J Hum Genet. 1999;64:706–711. doi: 10.1086/302296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss J, Gross-Hardt S, Christensen E, Schmidt P, Mendel RR, Schwarz G. A mutation in the gene for the neurotransmitter receptor-clustering protein gephyrin causes a novel form of molybdenum cofactor deficiency. Am J Hum Genet. 2001;68:208–213. doi: 10.1086/316941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria-Araujo JA, Fischer B, Otte T, Nimtz M, Mendel RR, Wray V, Schwarz G. The tetrahydropyranopterin structure of the sulfur-free and metal-free molybdenum cofactor precursor. J Biol Chem. 2004;279:15994–15999. doi: 10.1074/jbc.M311815200. [DOI] [PubMed] [Google Scholar]
- Sass JO, Nakanishi T, Sato T, Shimizu A. New approaches towards laboratory diagnosis of isolated sulphite oxidase deficiency. Ann Clin Biochem. 2004;41:157–159. doi: 10.1258/000456304322880078. [DOI] [PubMed] [Google Scholar]
- Schwahn BC, Galloway PG, Bowhay S, Veldman A, Santamaria JA, Schwarz G, Belaidi AA. Successful treatment of two neonates with molybdenum cofactor deficiency (MOCD) type a, using cyclic pyranopterine monophosphate (CPMP) J Inherit Metab Dis. 2010;33:S29. [Google Scholar]
- Schwarz G, Santamaria-Araujo JA, Wolf S, Lee HJ, Adham IM, Grone HJ, Schwegler H, Sass JO, Otte T, Hanzelmann P, Mendel RR, Engel W, Reiss J. Rescue of lethal molybdenum cofactor deficiency by a biosynthetic precursor from Escherichia coli. Hum Mol Genet. 2004;13:1249–1255. doi: 10.1093/hmg/ddh136. [DOI] [PubMed] [Google Scholar]
- Schwarz G, Mendel RR, Ribbe MW. Molybdenum cofactors, enzymes and pathways. Nature. 2009;460:839–847. doi: 10.1038/nature08302. [DOI] [PubMed] [Google Scholar]
- Sie SD, de Jonge RC, Blom HJ, Mulder MF, Reiss J, Vermeulen RJ, Peeters-Scholte CM (2010) Chronological changes of the amplitude-integrated EEG in a neonate with molybdenum cofactor deficiency. J Inherit Metab Dis DOI: 10.1007/s10545-010-9198-z (Epub ahead of print). [DOI] [PMC free article] [PubMed]
- Tan WH, Eichler FS, Hoda S, Lee MS, Baris H, Hanley CA, Grant PE, Krishnamoorthy KS, Shih VE. Isolated sulfite oxidase deficiency: a case report with a novel mutation and review of the literature. Pediatrics. 2005;116:757–766. doi: 10.1542/peds.2004-1897. [DOI] [PubMed] [Google Scholar]
- Vasiliades J. Reaction of alkaline sodium picrate with creatinine: I. Kinetics and mechanism of formation of the mono-creatinine picric acid complex. Clin Chem. 1976;22:1664–1671. [PubMed] [Google Scholar]
- Veldman A, Santamaria-Araujo JA, Sollazzo S, Pitt J, Gianello R, Yaplito-Lee J, Wong F, Ramsden CA, Reiss J, Cook I, Fairweather J, Schwarz G. Successful treatment of molybdenum cofactor deficiency type A with cPMP. Pediatrics. 2010;125:e1249–e1254. doi: 10.1542/peds.2009-2192. [DOI] [PubMed] [Google Scholar]
- Wadman SK, Cats BP, Debree PK. Sulfite oxidase deficiency and the detection of urinary sulfite. Eur J Pediatr. 1983;141:62–63. doi: 10.1007/BF00445675. [DOI] [Google Scholar]
- Wahl B, Reichmann D, Niks D, Krompholz N, Havemeyer A, Clement B, Messerschmidt T, Rothkegel M, Biester H, Hille R, Mendel RR, Bittner F. Biochemical and spectroscopic characterization of the human mitochondrial amidoxime reducing components hmARC-1 and hmARC-2 suggests the existence of a new molybdenum enzyme family in eukaryotes. J Biol Chem. 2010;285:37847–37859. doi: 10.1074/jbc.M110.169532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Vincent AS, Halliwell B, Wong KP. A mechanism of sulfite neurotoxicity: direct inhibition of glutamate dehydrogenase. J Biol Chem. 2004;279:43035–43045. doi: 10.1074/jbc.M402759200. [DOI] [PubMed] [Google Scholar]