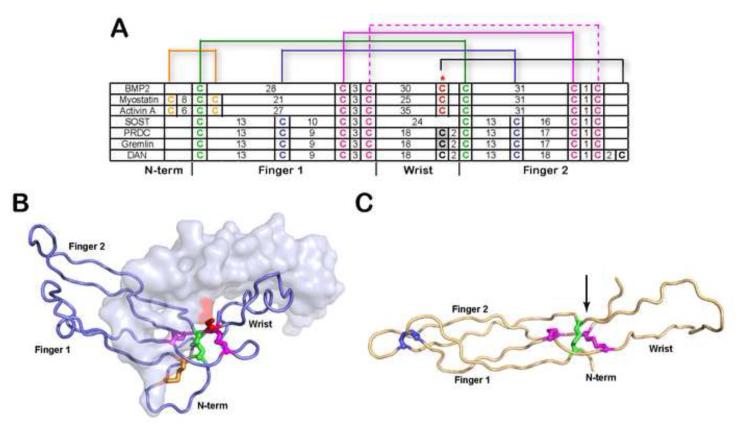
Figure 1. Comparison of cysteine organization of DAN family antagonists to TGF-β ligands.
A) Disulfide linkage comparison of representative TGF-β ligands (BMP2, myostatin and activin A) and DAN family of antagonists. The number of amino acids spanning consecutive cysteines is indicated. The red asterisk indicates the cysteine responsible for dimerization in TGF-β ligands. For PRDC and gremlin, the putative unpaired cysteine implicated in dimerization is highlighted gray. Brackets represent disulfide-linkages. B) X-ray structure of myostatin (PDB:3HH2) depicting one monomer in surface and one in ribbon and C) NMR structure of SOST (PDB:2K8P). Disulfide bonds in (A-C) are colored consistently. The cystine knot is shown where two disulfide bonds on opposing β-strands (purple) form a ring structure with the polypetide chain linked through the center of the ring by a single disulfide bond (green). The arrow in (C) indicates the position of the unpaired cysteine in PRDC through alignment with SOST.