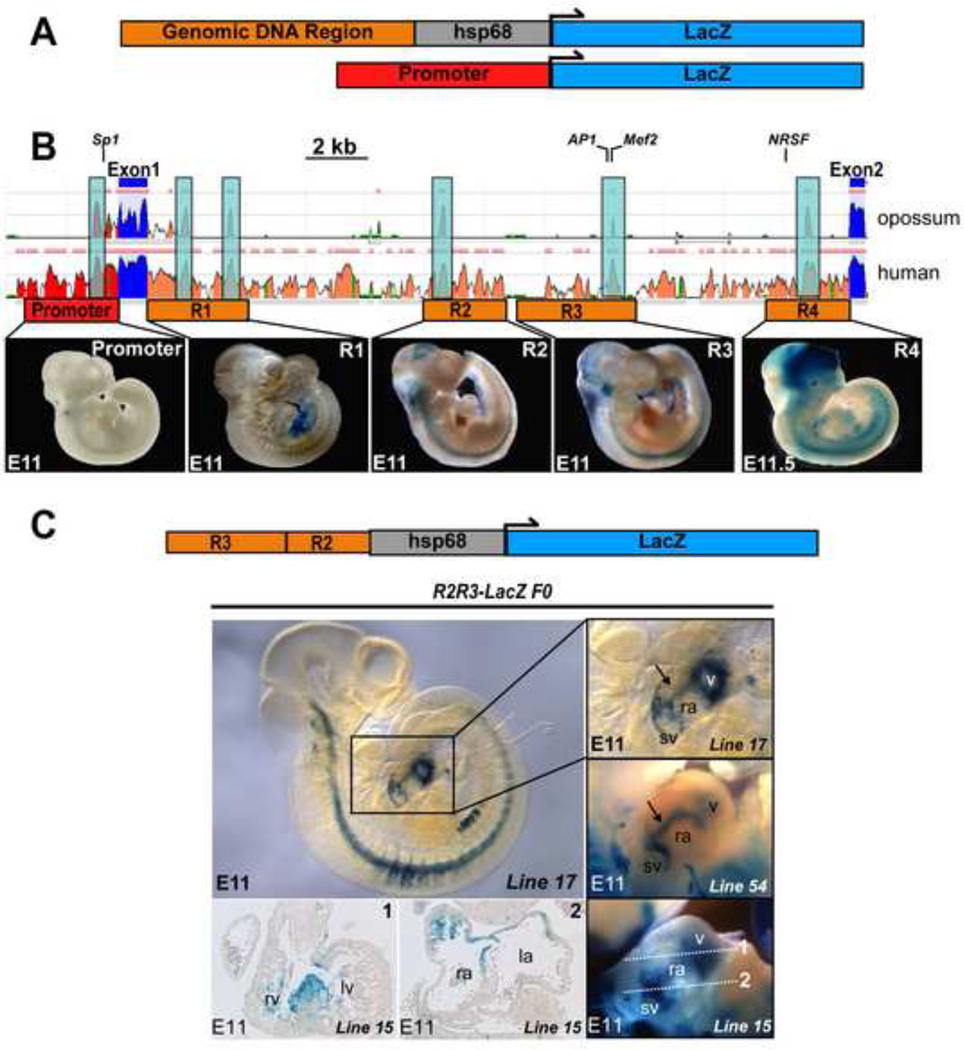
Figure 2. Identification and in vivo testing of Hcn4 regulatory elements.
(A) Genomic regions containing putative CREs were cloned upstream of hsp68LacZ or LacZ (basal promoter) to generate constructs for pronuclear injection. (B) Three species alignment at the Hcn4 locus from the ECR Browser, with mouse as the base genome against human and opossum. Peaks reflect degree of conservation (scale: 50–100%). Color code: red, upstream non-coding DNA; salmon, intronic DNA; blue, exons. Multiple conserved elements were selected for analysis (1–4) from Intron 1. Locations of previously validated transcription factor binding sites (TFBS) are indicated (NRSF, Mef2, AP-1, Sp1). Examples of founders from each construct injected are shown below the corresponding area on the Hcn4 genomic map. Although the presence of variable extra-cardiac reporter activity among different founders confirmed reporter insertion into permissive loci, none of the founders exhibited consistent reporter activity within the heart. (C) Left, an E11 embryo from an R2R3-LacZ transgenic line (line 17) stained with bluo-gal reporter activity within defined areas of the heart. Right, the cardiac region of line 17 in comparison with embryos from two additional transgenic founders (line 15 and line 22) shows similar cardiac reporter activity despite different genomic integration sites. By whole mount imaging, activity is concentrated at the junction of the sinus venosus and the right atrium, a linear area extending from the sinus venosus at the level of the interatrial groove to the ventricles (arrow), and within the ventricles. Sections from one founder embryo (line 15, bottom left) show reporter activity in the interventricular ring at the crest of the developing interventricular septum (section 1); and between the sinus venosus and atrium (section 2). Abbreviations: sv, sinus venosus; ra, right atrium; la, left atrium; v, ventricles; rv, right ventricle; lv, left ventricle.