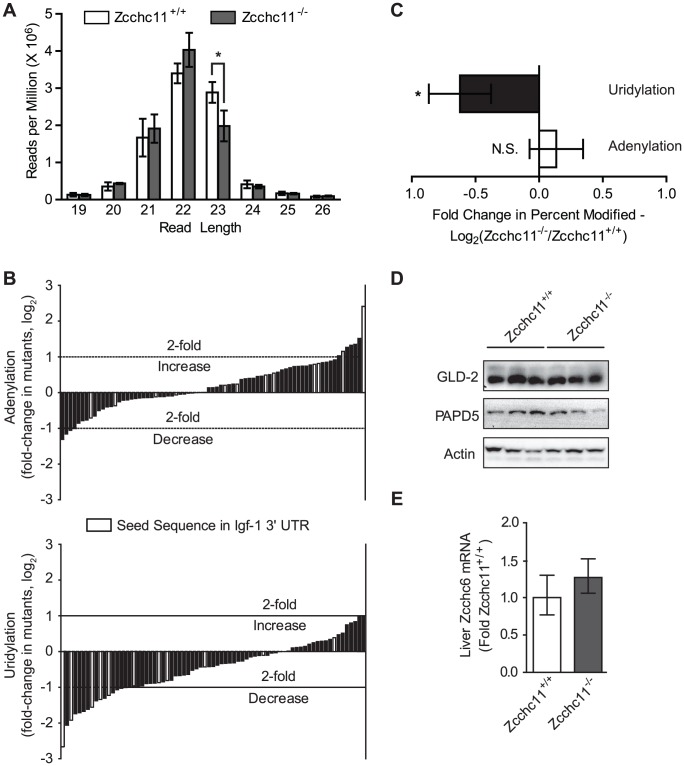
Figure 3. Zcchc11 is essential for the uridylation of miRNAs.
The miRNA libraries from Zcchc11-defficent and wild type livers at 8 days of age were analyzed for differences in sequence diversity. (A) Histogram of the number of sequences of a given read length across the libraries, showing fewer 23 nucleotide-long reads in the mutant samples. *p<0.05 by two-way ANOVA with Bonferroni post hoc test. (B) End modifications, identified as any nucleotide one base beyond the length of the miRNA published in miRBase, were quantified for each library. Most miRNAs ending in uridine in the control mice did so less frequently in Zcchc11-deficient mice. The mean of the fold change in the percent of sequences adenylated or uridylated between the knockout and wild type were graphed as waterfall plots, with every bar representing the geometric mean of (percent of miR-X sequences modified in the mutants)/(percent of miR-X sequences modified in the WT) for the three separate libraries. Only miRNAs with >10,000 reads and >0.1% sequences end-modified in the wild type libraries were included. Data were expressed as fold-change in Zcchc11−/− mice compared to Zcchc11+/+ mice. Light grey bars indicate miRNAs with a seed sequence complementary to a portion of the IGF-1 3′ UTR. (C) Average fold change across all miRNA species shown in Figure 3B, showing that uridines but not adenines were less frequent at miRNA termini due to Zcchc11 deficiency. Error bars indicate 95% c.i. *p<0.05 vs. 0 by student's t-test. (D) Immunoblots for the indicated non-canonical poly(A) polymerases in tissue homogenates prepared from the livers of 8 day-old mice, showing no effect of genotype. (E) qRT-PCR for Zcchc6 expression in the livers of 8 day-old mice, showing no effect of genotype.