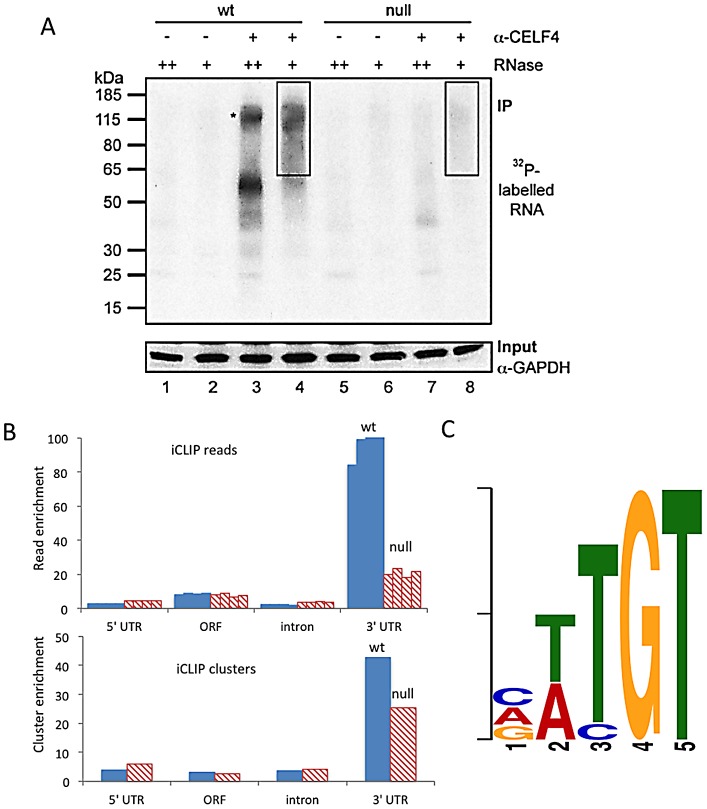
Figure 1. CELF4 binds mRNAs mostly in the 3′ UTR and favors an (A/U)UGU binding motif.
A. Rigorous purification of CELF4-bound RNAs with iCLIP. The autoradiogram shows size-separated crosslinked protein-RNA complexes following complete digestion with high (++) or partial digestion with low (+) amounts of RNase I, immunopurification with an anti-CELF4 antibody and 5′ end radiolabeling. The boxes depict the areas on the nitrocellulose membrane from which crosslinked RNAs were purified for reverse transcription. The asterisk marks dimerized CELF4. B. Gene segment analysis showing 3′ UTR enrichment of CELF4 binding. For enrichment, the percentage of reads (upper panel) or clusters (lower panel) mapping to a particular gene segment is divided by the percentage of the genome encoding this type of segment. For read enrichment, individual replicates are shown, and for cluster enrichment, the wildtype and knockdown experiments were grouped. C. The most significant CELF4 regulatory motif discovered by comparison of significant crosslink clusters determined by CELF4 iCLIP from wildtype or Celf4 null brain. The e-value of the top motif, as determined by DREME software, was 4.7×10−338.