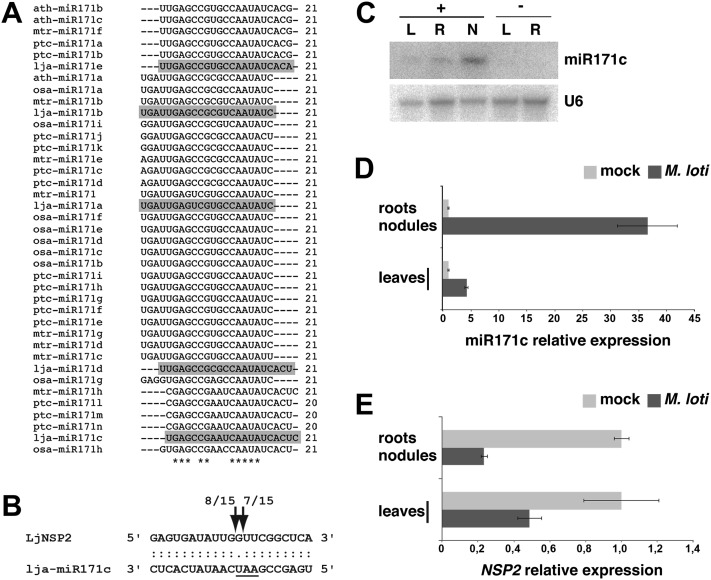
Figure 5.
Lja-miR171c targets NSP2 and is up-regulated in L. japonicus nodule tissue. A, Alignment of mature miR171 isoforms from L. japonicus and several plant species. L. japonicus miR171 members, marked in gray, were identified in this study by cloning (lja-miR171b to lja-miR171e) or homology mapping (lja-miR171a). Rice, poplar, Arabidopsis, and M. truncatula sequences were retrieved from the miRBase registry. Sequences were aligned using Clustal version 2.1; ath, Arabidopsis; lja, L. japonicus; mtr, M. truncatula; osa, rice; ptc, poplar. B, Validation of NSP2 as a miR171c target in L. japonicus. Cleavage sites were determined by 5′ RACE analysis. The number of sequenced fragments ending at a respective site (out of total clones sequenced) is indicated. Predicted sites of endonucleolytic cleavage are indicated by arrows. The nucleotide triplet that is different from the canonical miR171 family members is underlined. C, miR171c accumulation in L. japonicus leaves (L), roots (R), and nodules (N) from M. loti-inoculated (+) or mock-treated (−) plants. Tissues were collected at 21 dpi. Root (R) samples from M. loti-inoculated plants (+) comprise remaining root tissue after nodule excision. D and E, Relative expression levels of mature miR171c (D) and NSP2 (E) in nodules compared with mock-treated roots and leaves from inoculated compared with mock-treated plants. Light gray bars indicate tissues harvested from mock-treated plants, and dark gray bars indicate tissues harvested from inoculated plants. All tissues were harvested at 21 dpi. Three biological and two technical replicates were analyzed. Error bars show the se.