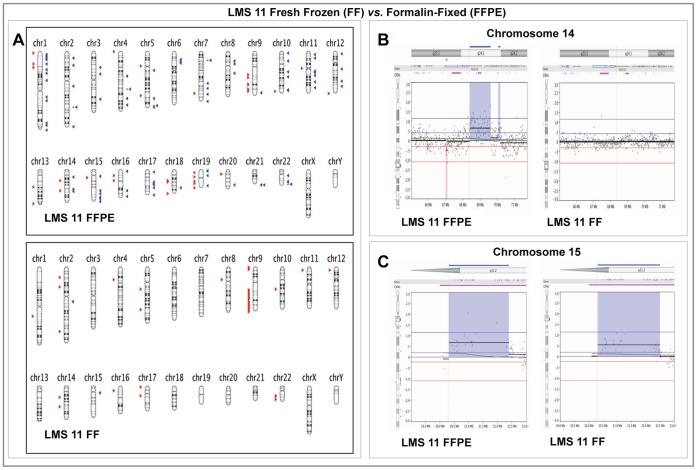
Figure 4. Comparison of Array CGH results in paired Fresh Frozen and Formalin-fixed Paraffin -Embedded samples from LMS 11.
Panel A: Graphical whole-genome views of both sample types showing that majority of the copy number aberrations (CNAs) identified in the macro-dissected FFPE sample were not detected in the FF sample. Deletions on the long arms of chromosomes 9, 14 and 15 as well as the short arm of chromosome 16 were the called on both sample types. Panel B: High resolution graphical views of a 6 Mb region along on Chromosome 14 (14q24.1) showing a group of probes with an average log2 ratio of approximately 0.6 and the corresponding single copy amplification detected in the FFPE sample but no aberrations detected in the FF sample. Panel C: High-resolution graphical views showing a closely similar copy number aberration detected on Chromosome 15 (15q11.2) in both sample types with similar probe log2 ratios. On Panel A, aberrations called by FASST2 algorithm are represented by blue triangles to the right (amplifications) and red triangles to the left (deletions) of the chromosomes. Double blue and red triangles/lines represent high-level amplifications and two-copy deletion, respectively. On Panels B and C, dots represent individual probe log2 ratios plotted as a function of their chromosomal position with a moving average of probe log2 ratios (wavy dark blue line). Aberration calls are represented by thick black lines with corresponding shaded blue areas above (amplifications) and red areas below (deletions) the zero line.