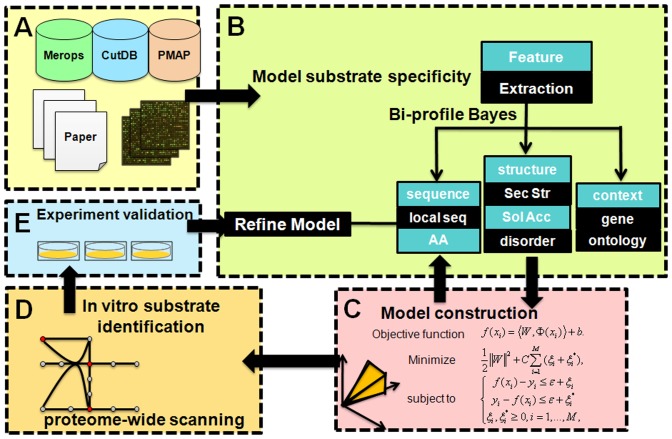
Figure 1. Schematic overview of the PROSPER approach.
There are several stages: (A) training datasets and independent test dataset of protease substrates were extracted from multiple resources. These included major comprehensive databases such as MEROPS, CutDB and PMAP, as well as recent proteome-wide profiling studies or the literature. (B) Useful sequence and structure features flanking the cleavage sites were derived and investigated, including local amino acid sequences, predicted secondary structure, solvent accessibility and native disorder. (C) The derived sequence and structural features were entered, following which cleavage probability models were built based on support vector regression (SVR) from the training dataset. In particular, the bi-profile Bayesian feature extraction was applied to extract and integrate the derived features into SVR models, which have been shown to be able to further improve prediction performance. (D) After building the PROSPER models, substrate sequence scanning predictions were made, and (E) PROSPER was further validated using a set of recently identified novel substrates reported in the literature or experimentally verified using positional proteomic approaches.