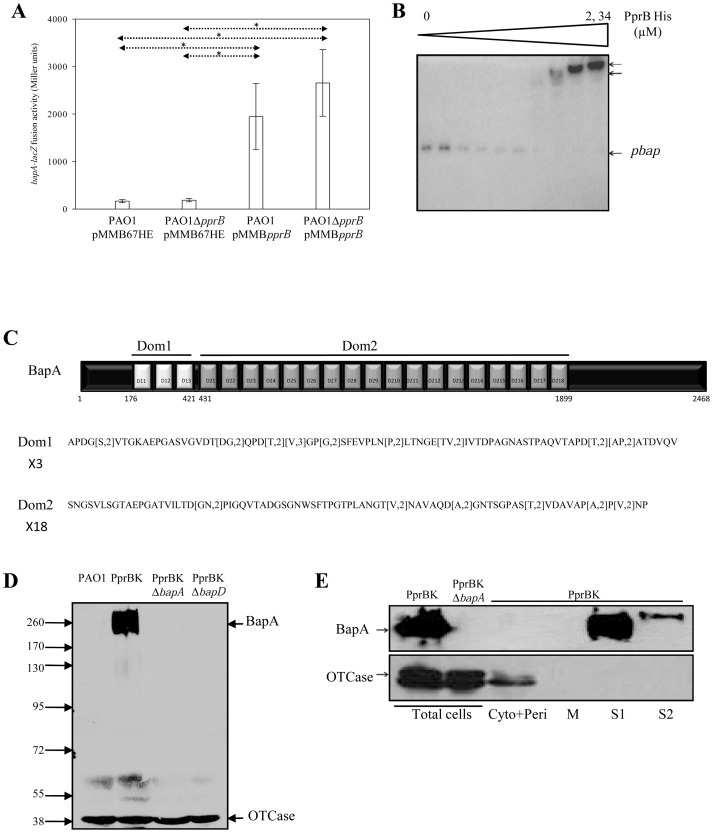
Figure 2. Validation of bap genes.
Expression of the chromosomal bapA-lacZ fusion (A) was monitored in the PAO1/pMMB67HE, PAO1ΔpprB/pMMB67HE, PAO1/pMMBpprB and PAO1ΔpprB/pMMBpprB strains. Data are expressed in Miller units and correspond to mean values (with error bars) obtained from three independent experiments. Statistical analysis was performed using a Welch test (*:<0.05). EMSA (B) performed with the purified PprB-6His protein, at concentrations of 0 to 2.34 µM, and the putative bap promoter DNA region. Two retarded complexes (upper arrows) were identified at high PprB-6His concentrations. BapA protein from P. aeruginosa is a 2468 aa polypeptide (C) organized into two domains of 3 (Domain 1) and 18 (Domain 2) repeats of 82 and 86 aa, respectively. BapA production was detected (D) in western blot in the total cell extracts obtained from PAO1, PprBK, PprBKΔbapA, and PprBKΔbapD strains. As a control of equivalent loading between strains, OTCase protein was detected in the same samples. Number on left side is molecular weight standard (kDa). BapA localization (E) was checked in soluble (cytoplasmic and periplasmic fractions), in membrane (M) fractions, in classical (S1) and associated loosely with the cell surface (S2) supernatants. Cell leakage was specifically detected using anti-OTCase antibody.