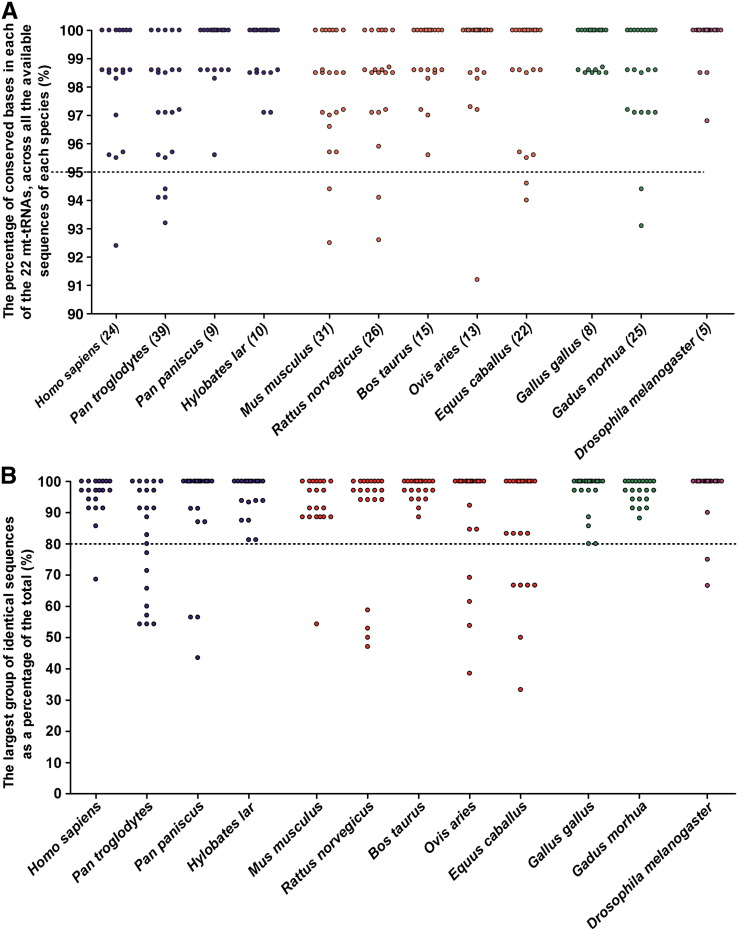
Fig. 3.
Conservation of the mt-tRNAs. A) This graph shows the percentage of bases within each mt-tRNA that are completely conserved across each sequence analysed for each species. Each mt-tRNA is represented by a dot and the total number of bases showing variation across the 22 mt-tRNAs in each species is displayed in the x-axis. B) Each point represents the percentage of identical sequences that make up the largest group for each mt-tRNA. The mt-tRNA identity of each individual point on either graph can be determined using Supplementary Fig. 2. In both Section A and Section B, the datda is split into different coloured groups (from left to right): primates (blue), other mammals (red), other vertebrates (green) and other metazonas (purple).
Conservation of the mt-tRNAs. A) This graph shows the percentage of bases within each mt-tRNA that are completely conserved across each sequence analysed for each species. Each mt-tRNA is represented by a dot and the total number of bases showing variation across the 22 mt-tRNAs in each species is displayed in the x-axis. B) Each point represents the percentage of identical sequences that make up the largest group for each mt-tRNA. The mt-tRNA identity of each individual point on either graph can be determined using Supplementary Fig. 2. In both Section A and Section B, the datda is split into different coloured groups (from left to right): primates (blue), other mammals (red), other vertebrates (green) and other metazonas (purple).