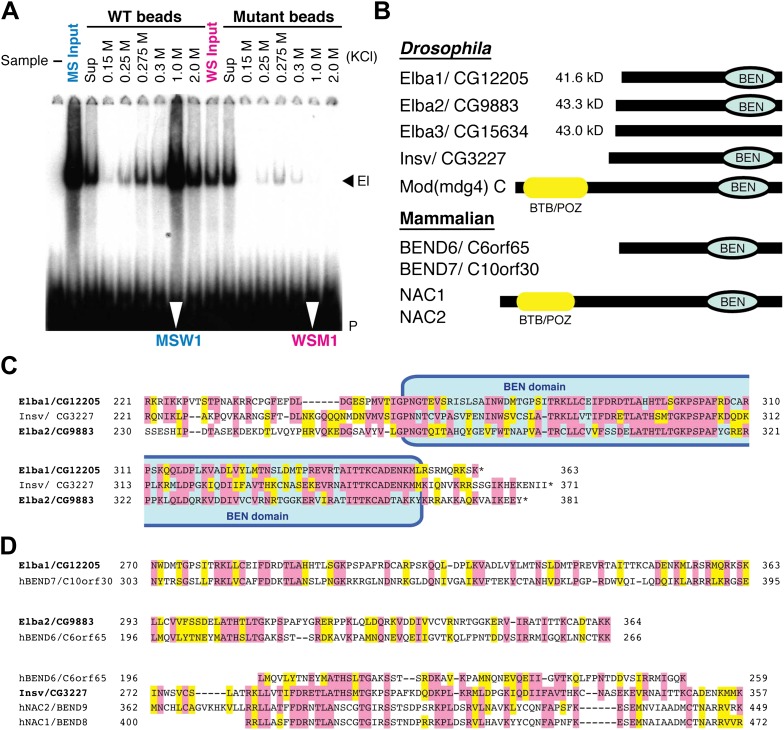
Figure 3. EMSA and Elba factor proteins.
(A) EMSA of fractions from the cross-affinity purification. The 32P-labeled Elba probe was incubated with fractions as indicated and subjected to 4% acrylamide-gel electrophoresis. El: Elba shift. P: probe. (B) Schematic of the Elba factors and BEN domain-containing (green) orthologs. BTB/POZ (yellow) domain is absent from Elba factors but is present in related proteins. (C) Sequence alignment of the C-terminal half of the Drosophila Elba1, Elba2 and a third Drosophila Ben protein Insv (Insensitive). The sequences of the three proteins were aligned according to the results of NCBI (National Center for Biotechnology Information) blast search (bl2seq). The amino acid residues conserved in more than two proteins are shaded with red. The residues that have similarities with each other are shaded with yellow. The predicted BEN domain region is boxed with pale blue. (D) Sequence alignment of Drosophila and mammalian (human) orthologs of BEN domain proteins. The C-terminal sequences of Elba1, Elba2 and Insv were subjected to blast search with human databases. Within the BEN domain sequences, the closest human ortholog of Elba1 is BEND7 (BEN domain-containing 7)/C10orf30 (29% identical, 46% positive), whereas for Elba2 the closest ortholog is BEND6/C6orf65 (29% identical, 53% positive). The BEN domain of Insv is most similar to three proteins: NAC2 (Nucleus accumbens-associated protein 2, NACC2)/BEND9, BEND6 and NAC1 (Nucleus accumbens-associated protein 1, NACC1)/BEND9. The amino acid residues conserved with each Drosophila protein are shaded with red. The residues that have similarities with each Drosophila protein are shaded with yellow.