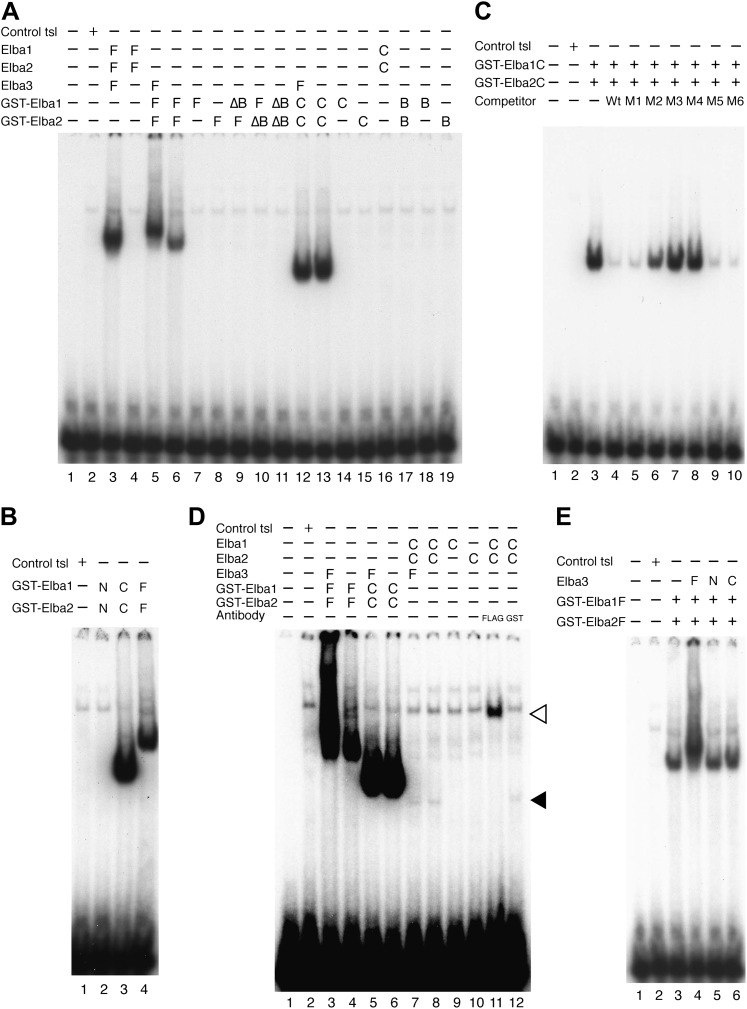
Figure 6. Characterization of Elba complex.
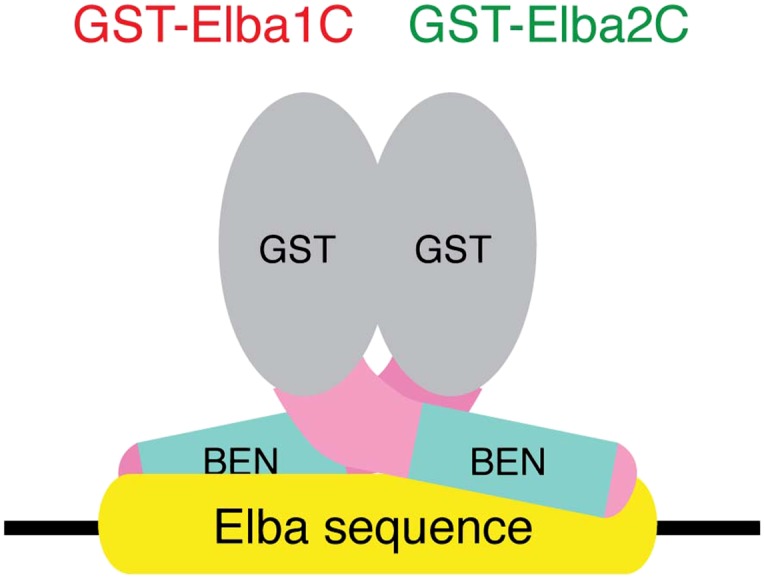
(A) DNA binding activity of different Elba protein variants. The proteins were translated in vitro from the mixed mRNAs shown above the lanes. All proteins used in this figure are FLAG tagged and approximately the same amounts of the translated proteins were added to each lane. The identities of the proteins added to each lane are indicated above the lane. For example, lane 3 has all three full length Elba proteins, while in lane 5 full length Elba3 is combined with full length Elba1 and Elba2 proteins that have an N-terminal GST tag. Lane 13 has GST fused to the C-terminal half of Elba1 and Elba2, while in lane 16 the C terminal halves of Elba1 and Elba2 lack the GST moiety. (B) GST-fused Elba1F:Elba2F (F) or Elba1C:Elba2C (C) binds to the Elba probe while Elba1N:Elba2N (N) does not. (C) Sequence specificity of the GST-Elba1C:GST-Elba2C dimer is the same as the nuclear/reconstituted hetero-tripartite complex (Figure 4). The EMSA experiment with GST-Elba1C:GST-Elba2C were performed in the absence (lanes 1–3) or presence of 100-fold excess of non-labeled competitors as indicated above the lanes. The sequences of competitor DNAs are shown in Figure 1. (D) Elba1C and Elba2C proteins lacking the N-terminal GST moiety have a weak DNA binding activity. Position of Elba1C:Elba2C shift is indicated by closed arrowhead. Position of the Elba1C:Elba2C FLAG supershift is indicated by open arrowhead. Proteins added to each lane including FLAG and GST antibodies are indicated above the lane. Note that the same shift was detected in lane 16 of (A) when the X-ray film was exposed for a longer period of time. (E) Supershifts of the GST-Elba1F:GST-Elba2F generated by the addition of Elba3. Proteins corresponding to the full length Elba3 (F), or the N-terminal (N) and C-terminal (C) halves of Elba3 were added to an EMSA reaction mix containing the GST-Elba1:GST-Elba2 dimer. Proposed structures of the native Elba complex, the artificial GST-dependent dimer by GST-Elba1:GST-Elba2 and GST-Elba1C:GST-Elba2C are shown in Figure 6—figure supplement 1–3, respectively.
Figure 6—figure supplement 1. A schematic model for the tripartite Elba complex.
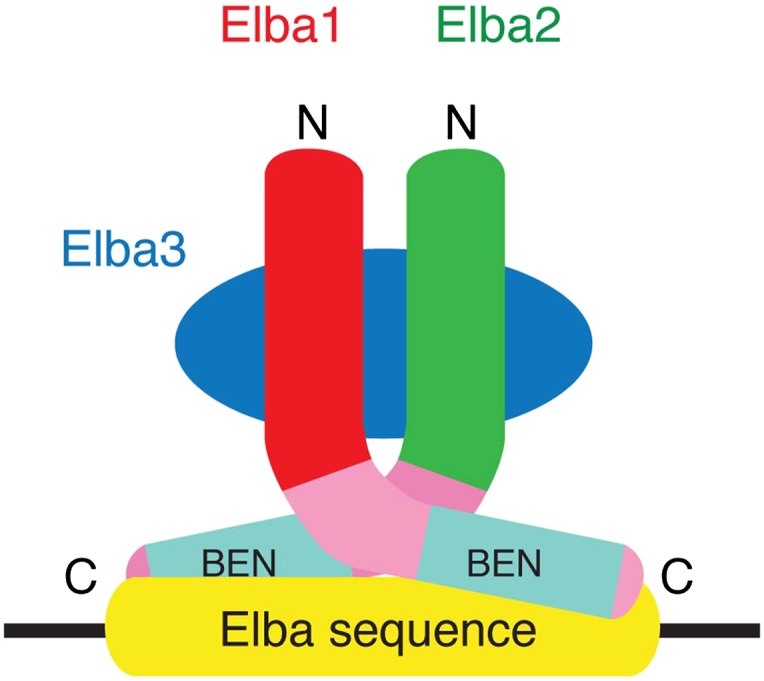
Figure 6—figure supplement 2. The artificial GST-Elba1:GST-Elba2 heterodimer binds to the asymmetric Elba recognition sequence.
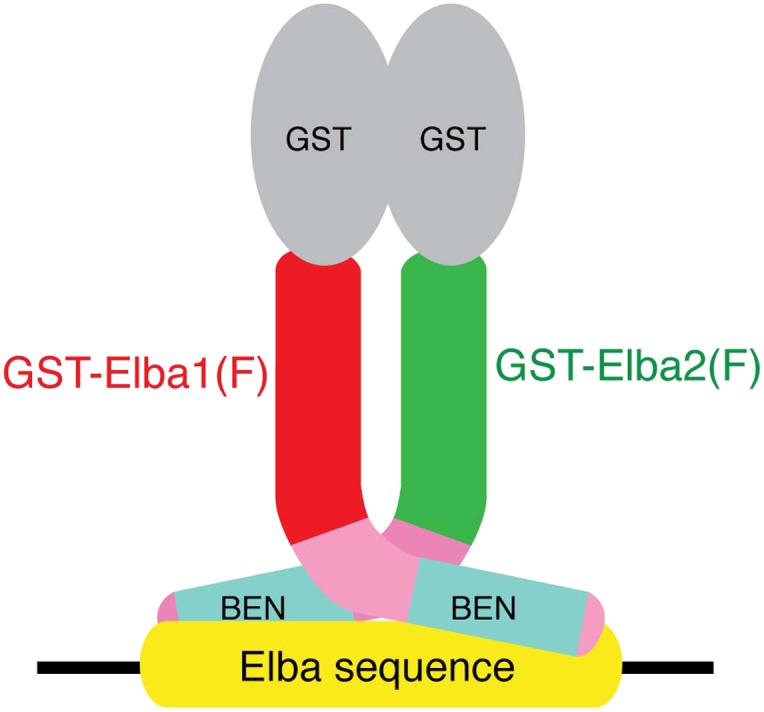
Figure 6—figure supplement 3. The artificial heterodimer of GST-Elba1C:GST-Elba2C binds to the asymmetric Elba recognition sequence.