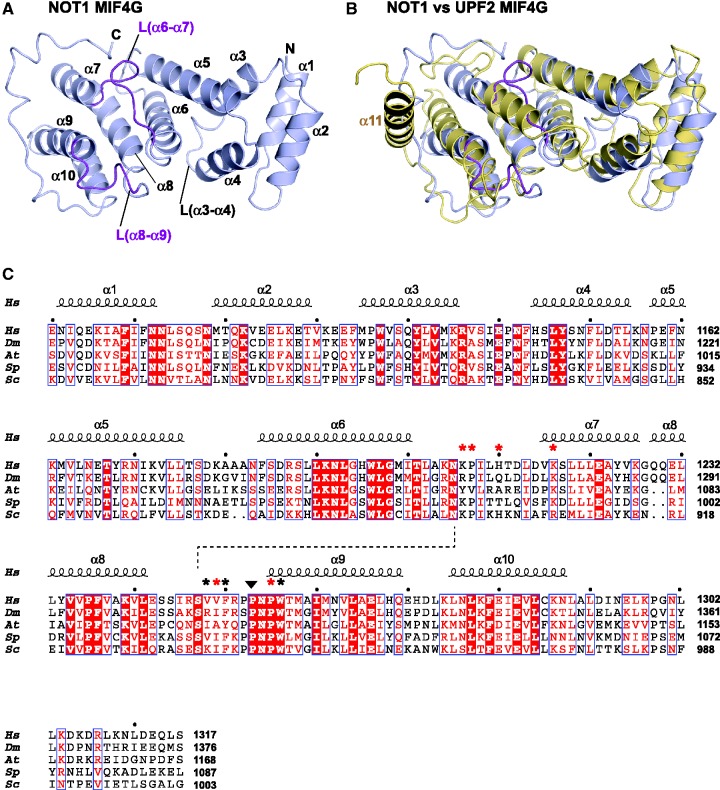
Figure 4.
Structure of the NOT1 MIF4G domain. (A) Cartoon diagram of the NOT1 MIF4G domain. Secondary structure elements are indicated. The CAF1-interacting parts of loops L(α6–α7) and L(α8–α9) are colored purple. (B) Comparison of NOT1 and UPF2 (PDB ID: 1UW4) MIF4G domains. NOT1 and UPF2 are colored light blue and yellow, respectively. (C) Structure-based sequence alignment of NOT1 MIF4G domains from various species. Species are as follows: Hs (Homo sapiens), Dm (D. melanogaster), At (Arabidopsis thaliana), Sp (S. pombe) and Sc (S. cerevisiae). Residues that are conserved in all aligned proteins are in red boxes, residues showing >70% similarity are printed in red, and the main residues involved in the interface are marked by asterisks (red if mutated in this study). The secondary structure elements derived from the structure of NOT1 are shown above the alignment and the conserved cis-proline is marked by a black triangle. The dashed line indicates a hydrogen bond between the invariant residues N1207 [L(α6–α7)] and S1249 [L(α8–α9)].