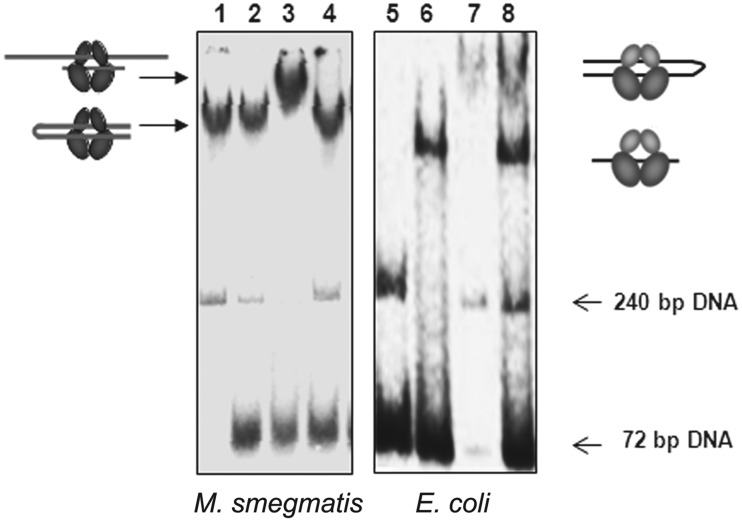
Figure 2.
EMSA of M. smegmatis and E. coli gyrase with 72- and 240-bp DNA. DNA gyrase (50 nM) was incubated with DNA and 2 µg/ml MFX at 37°C for 10 min and then shifted back to ice before adding the second DNA. The 72- and 240-bp DNA used were the same as in Figure 1. Lane 1—MsGyr + 240 bp, Lane 2—MsGyr + 240 bp→72 bp (where the arrow-head indicates order of addition), Lane 3—MsGyr + 72 bp+ MFX→240 bp, Lane 4—MsGyr + 240 bp+ MFX→72 bp, Lane 5—Free 72- and 240-bp DNA, Lane 6-EcGyr + 72 bp, Lane 7-EcGyr + 240 bp, Lane 8—EcGyr + 72 bp+ MFX→240 bp. The complexes were resolved on a 5% native polyacrylamide gel and visualized using a phosphor imager. The pictograms indicate the likely mode of binding of the DNAs to the gyrase holoenzymes.