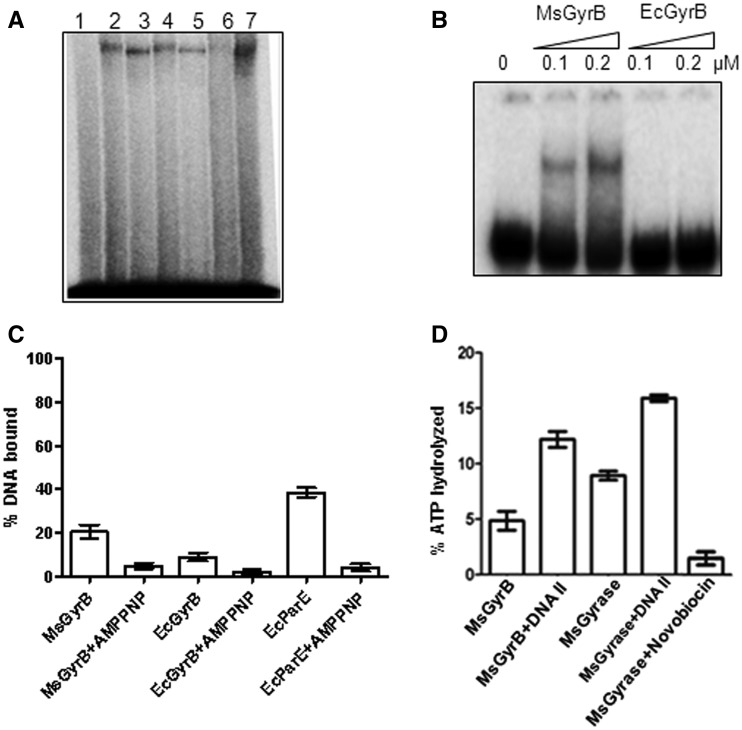
Figure 3.
DNA binding by GyrB. (A) Crosslinking of DNA gyrase holoenzymes and individual subunits from M. smegmatis and E. coli with 72-bp DNA. An amount of 100 nM of GyrA subunit and 1 µM of each of the GyrB subunits were used for individual subunit crosslinking. The holoenzymes were reconstituted by mixing the 100 nM of GyrA subunit with 200 nM of the respective GyrB subunits. DNA crosslinking with gyrase and its individual subunits was carried out as described in ‘Materials and Methods’ section and the products were resolved on an 8% SDS–PAGE. Lane 1—DNA only, Lane 2—EcGyr, Lane 3—MsGyr, Lane 4—EcGyrA, Lane 5—MsGyrA, Lane 6—EcGyrB and Lane 7—MsGyrB; (B) EMSA with 72-bp DNA using MsGyrB and EcGyrB (0.1 and 0.2 µM) subunits. The DNA-bound complexes were resolved on a 5% native polyacrylamide gel; (C) Filter-binding assay with 20 nM each of MsGyrB, EcGyrB and E. coli ParE (EcParE) using 72-bp DNA. Enzyme was pre-incubated with 2 mM AMPPNP before addition of DNA (where indicated); (D) DNA (72 bp)-mediated stimulation of ATPase activity. ATPase assays were carried out with MsGyr (100 nM) or its GyrB subunit (100 nM) in the absence and presence of 72-bp DNA (400 nM). Lane 1—MsGyrB alone, Lane 2—MsGyrB + DNA, Lane 3—MsGyr alone, Lane 4—MsGyr + DNA and Lane 5—MsGyr + Novobiocin (10 µg/ml). The error bars represent standard deviation across three measurements.